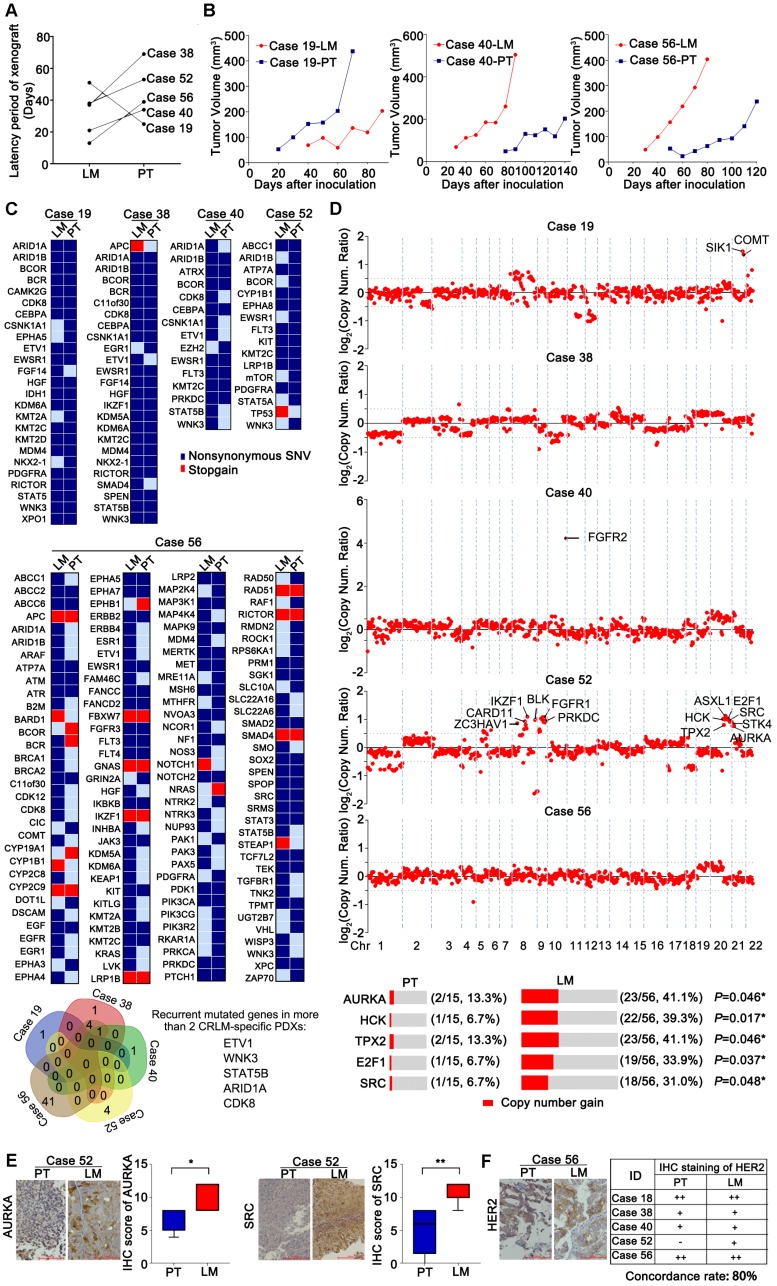
Figure 4.
Genomic alterations were compared between five pairs of PDX models from CRLM and CRPT, respectively. (A) The difference of latency period of xenografts between CRPT and CRLM PDXs. Four of five CRLM PDX models showed a faster growth than the corresponding CRPT PDXs, except for Case 19. (B) Representative growth curve of three paired PDXs (Cases 19, 40, and 56). (C) Numerous mutated genes were identified and compared in five paired PDX models. Several recurrent mutated genes in two more CRLM-specific PDXs were identified by Venn diagram in the bottom. Red, stopgain; Dark blue, nonsynonymous SNV. (D) Copy number alterations were also compared, displayed by log2 (copy number ratio). Values more than 0.5 was assumed to be different between primary tumors and corresponding liver metastases. Several copy number alterations of genes indicated in C1 were selected for the validation in a larger biobank of PDX models from CRLM/PT. Copy number ≥ 3 was considered as gain. (E) The expression of AURKA and SRC were further validated at protein level by IHC staining. The expression of AURKA and SRC in PDXs from CRLM were higher than that of PDXs from corresponding primary tumors. (F) HER2 expression was also evaluated in five paired PDX from CRLM. The table demonstrated an 80% overlap in HER2 expression. * p < 0.05; ** p < 0.01 by one-way ANOVA or chi-squared test.