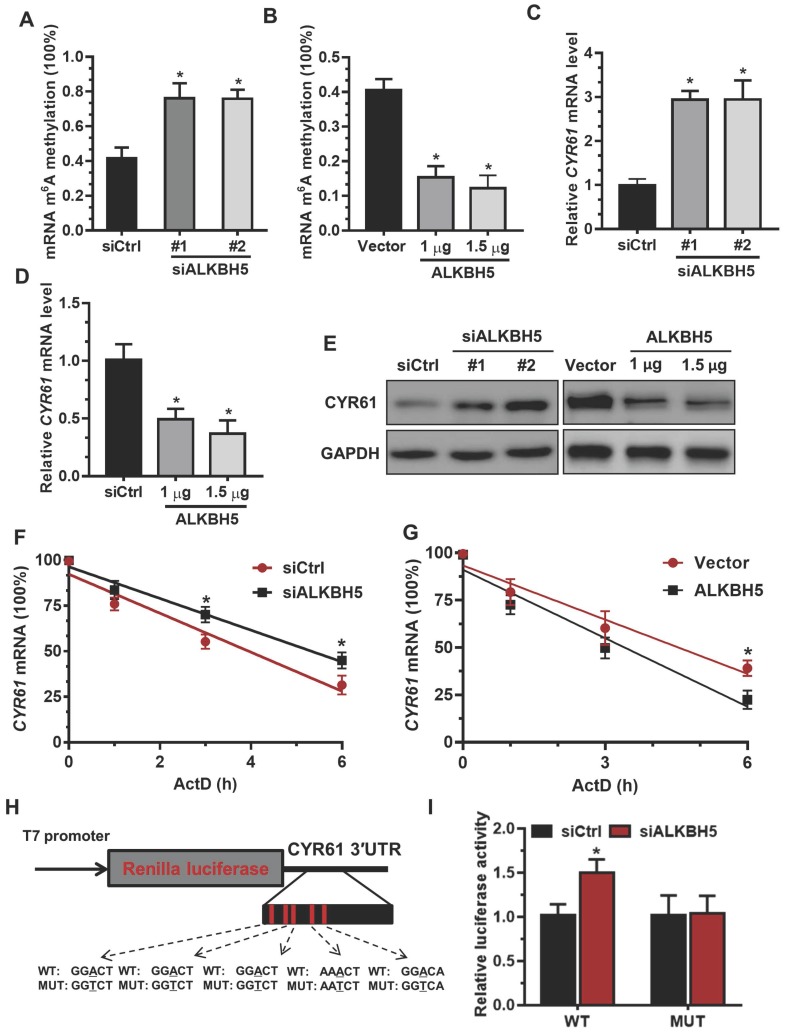
Figure 4.
The m6A mark is critical for CYR61 expression and stability in trophoblasts. (A and B) RNA m6A RNA methylation of primary trophoblast transfected with siALKBH5 or ALKBH5-expressing plasmid was assessed using the m6A RNA Methylation Quantification ELISA kit. (C-E) Western blotting and qRT-PCR analysis of CYR61 expression in HTR-8 cells transfected with siCtrl, siALKBH5 #1, siALKBH5 #2, control vector, or ALKBH5-expressing plasmid after 48 h. (F and G) HTR-8 cells were transfected with siCtrl, siALKBH5, vector or ALKBH5-expressing plasmid for 36 h; a time course for mRNA stability was initiated by adding an RNA-Polymerase II inhibitor [actinomycin D (5 µg/mL)]. Cells were harvested at the indicated time points. Expression levels were normalized to that at “0 hour”, and GAPDH mRNA was used as the reference gene. The results are shown as the mean of at least three independent experiments. *P < 0.05 versus the vector or siCtrl. (H and I) A schema for the constructs and co-transfection experiments, a fragment of 3'-UTR of CYR61 (wild-type and mutant) was cloned into a psiCheck2 vector, downstream of the renilla luciferase gene. These construct where was co-transfected with the siALKBH5 or siCtrl into HTR-8 cells. Cells were harvested after 24 h. Renilla luciferase activity was measured and normalized to that of firefly luciferase. The results are represented as the mean of three independent experiments. *P < 0.05 versus the siCtrl.