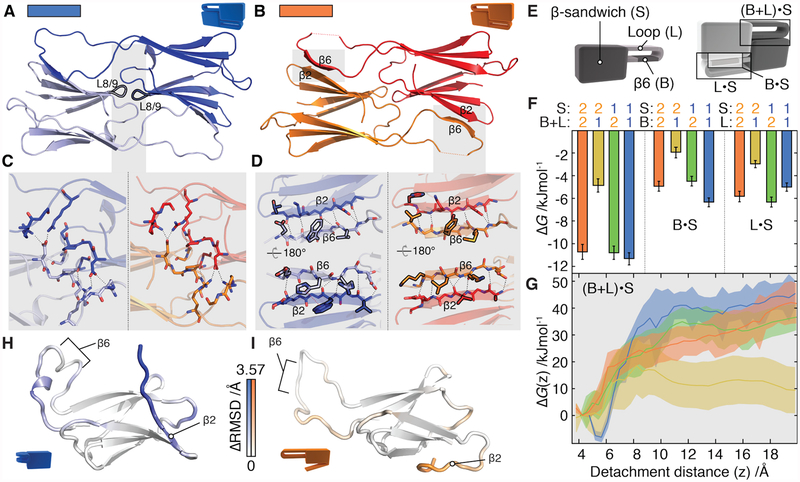
Figure 3. Selectivity in the structurally conserved α-crystallin domain.
A and B) α1 and α2 dimers have an identical fold (backbone RMSD = 1.2 Å) in which two highly similar interfaces (labelled L8/9 and β6· β2) connect monomers.
C) The L8/9 interface is centred on the loop between β8 and β9 (black outline) and is indistinguishable in the two proteins. Inter-chain hydrogen bonds are shown as dashed lines.
D) The two β6·β2 interfaces in the dimer are formed by exchange between the β6 and β2 strands. Side-chains that differ between α1 and α2 at homologous positions are outlined in black. The π-stacking interaction specific to α1 is shown as a dotted red line.
E) Constructs were designed by swapping the β-sandwich, loop, and β6 strand (left). These were used to assess the strength of the β6· β2 interface, and deconvolve the contribution from the loop and β6 strand (right).
F) Global thermodynamic model of dimerization based on experimentally determined ΔGα.α values in Fig S12G. The combined loop and β6 from α1 interact less favourably with β2 from α2 than all other combinations (left). α2 and α1 partition contributions to ΔGα.α differently (shaded). Error bars are standard deviations from 1000 bootstrap replicates of the model fit.
G) In a simulated heterodimer, the free energy barrier is significantly reduced for the α2· α1 pair (yellow), but indistinguishable from the homodimers in the case of α1·α2 (green) when the β6·β2 interface is disrupted along a reaction coordinate that separates them. Shaded area corresponds to the standard error of the mean.
H,I) Median monomeric conformations determined by principal component analysis coloured according to structural difference. This is calculated at each residue from the Cα RMSD between α1 and α2 monomers, minus the RMSD between repeats for each monomer. Positive ΔRMSD values indicate conformational differences between proteins that cannot be explained by the variations intrinsic to each protein, and only those with p<0.05 (after Bonferroni correction, permutation test) are coloured. Differences are apparent in the loop surrounding β6 and in β2. In α1 the loop curls up, whereas in α2 the β2 strand detaches readily from the remainder of the β-sandwich.