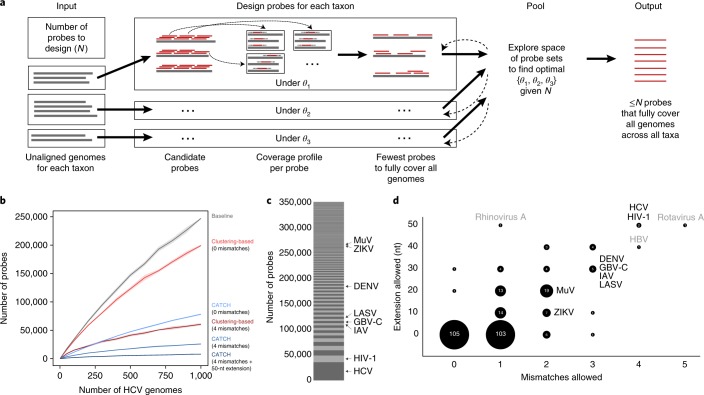
Fig. 1. Using CATCH for probe set design.
a, Sketch of CATCH’s approach to probe design, shown with three datasets (typically, each is a taxon). For each dataset d, CATCH generates candidate probes by tiling across input genomes and, optionally, reduces the number of them using locality-sensitive hashing. Then it determines a profile of where each candidate probe will hybridize (the genomes and regions within them) under a model with parameters θd (see Supplementary Fig. 1b for details). Using these coverage profiles, it approximates the smallest collection of probes that fully captures all input genomes (described in the text as s(d, θd)). Given a constraint on the total number of probes (N) and a loss function over θd, it searches for the optimal θd for all d. b, Number of probes required to fully capture increasing numbers of HCV genomes. Approaches shown are simple tiling (gray), a clustering-based approach at two levels of stringency (red), and CATCH with three choices of parameter values specifying varying levels of stringency (blue). See Supplementary Note 2 for details regarding parameter choices. Previous approaches for targeting viral diversity use clustering in probe set design. The shaded regions around each line are 95% pointwise confidence bands calculated across randomly sampled input genomes. c, Number of probes designed by CATCH for each dataset (of 296 datasets in total) among all 349,998 probes in the VALL probe set. Species incorporated in our sample testing are labeled. d, Values of the two parameters selected by CATCH for each dataset in the design of VALL: number of mismatches to tolerate in hybridization and length of the target fragment (in nucleotides) on each side of the hybridized region assumed to be captured along with the hybridized region (cover extension). The label and size of each bubble indicate the number of datasets that were assigned a particular combination of values. Species included in our sample testing are labeled in black, and outlier species not included in our testing are in gray. In general, more diverse viruses (for example, HCV and HIV-1) are assigned more relaxed parameter values (here, high values) than less diverse viruses, but still require a relatively large number of probes in the design to cover known diversity (see c). Panels similar to c and d for the design of VWAFR are in Supplementary Fig. 3.