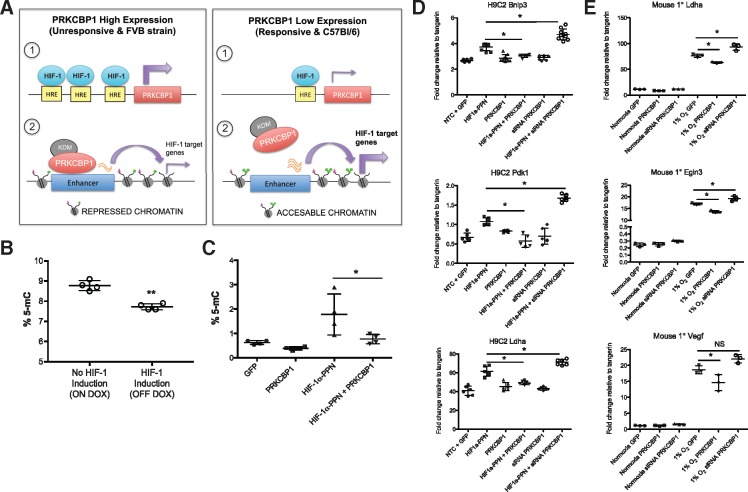
Figure 7.
Proposed mechanism. (A) Proposed mechanism of HIF-induced PRKCBP1 expression, and PRKCBP1 mediated negative feedback of HIF-1 regulated genes. (B) Global DNA methylation is altered following cardiac HIF-1 induction in mice. After 3 days of HIF-1 induction, ELISA analysis confirmed significant reduction in the number of methylated CG residues compared to HIF-1 non-induced mice. n = 4 per group, **P = 0.0007. (C) DNA methylation is altered in response to HIF-1 and PRKCBP1 in HL-1 cells. ELISA analysis confirmed significant increase in the number of methylated CG residues in HL-1 HIF-1α-PPN transfected cells, and co-transfection with PRKCBP1 reduced % 5-mC to baseline levels. n = 4 per group, *P < 0.05. PRKCBP1 modulates the HIF response in (D) H9C2 cells and (E) mouse primary cardiomyocytes. Overexpression of PRKCBP1 in the presence of (D) HIF-1α-PPN or (E) 1% O2 reduces HIF-1 target gene transcript. siRNA mediated knockdown of PRKCBP1, in the presence of HIF-1α-PPN or hypoxia removed this negative inhibition (n = 6 H9C2; n = 3 mouse primary cells). Two-tailed Student’s t-test (B) and one-way ANOVA with Bonferroni post hoc test (C–E) was used for statistical analysis. Data are expressed as mean ± SD. *P < 0.05. Adapted from Shen et al. HRE, hypoxia response element; KDM, lysine demethylase.