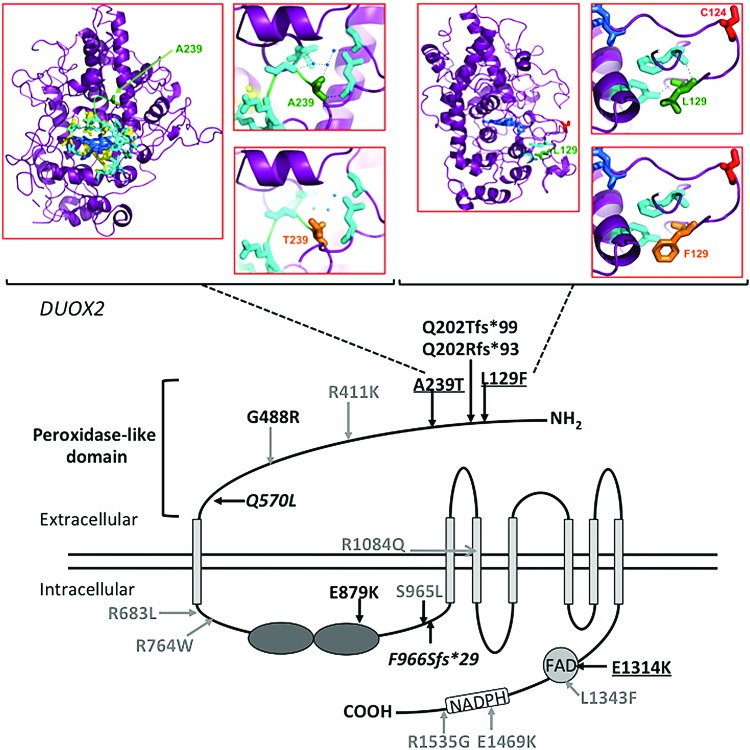
FIG. 2.
Schematic showing DUOX2, functional domains, and the position of mutations identified in this study. Known CH-associated mutations that have not undergone functional characterization are shown in gray. Known CH-associated truncating mutations or missense mutations shown to be pathogenic in vitro are shown in black. Novel mutations are shown in black and underlined. Recurrent mutations affecting more than two cases in this study are shown in italics. The model was constructed using data from the InterPro database. Homology modeling has been used to predict the consequences of the novel mutations A239T and L129F. A239 (green) is small and apolar, while T (orange) is large and polar and disrupts local polar contacts (blue) and spacefill, probably disrupting backbone H-bonds between A239 and H20. L129 (green) and F (orange) are both hydrophobic, but F is significantly larger and does not fit in the position of L129 that will affect the local structure. C124 (red) is situated in a loop that may be repositioned with the L129F mutation, and since C124 is reported to form a disulfide bridge with C1162, this may destabilize DUOX2. Color images are available online.