Abstract
Wilms tumour is a paediatric malignancy with features of halted kidney development. Here, we demonstrate that the Iroquois homeobox genes IRX3 and IRX5 are essential for mammalian nephrogenesis and govern the differentiation of Wilms tumour. Knock‐out Irx3 − /Irx5 − mice showed a strongly reduced embryonic nephron formation. In human foetal kidney and Wilms tumour, IRX5 expression was already activated in early proliferative blastema, whereas IRX3 protein levels peaked at tubular differentiation. Accordingly, an orthotopic xenograft mouse model of Wilms tumour showed that IRX3 −/− cells formed bulky renal tumours dominated by immature mesenchyme and active canonical WNT/β‐catenin‐signalling. In contrast, IRX5 −/− cells displayed activation of Hippo and non‐canonical WNT‐signalling and generated small tumours with abundant tubulogenesis. Our findings suggest that promotion of IRX3 signalling or inhibition of IRX5 signalling could be a route towards differentiation therapy for Wilms tumour, in which WNT5A is a candidate molecule for enforced tubular maturation. © 2018 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: Wilms tumour, Iroquois, IRX3, IRX5, nephrogenesis, differentiation, WNT5A
Introduction
Wilms tumour is the most common renal neoplasm in children. Its biology and morphology closely mirror the early stages of mammalian kidney development (nephrogenesis). A key element of nephrogenesis is the transformation of the blastemal elements of the metanephric mesenchyme, via mesenchymal‐to‐epithelial transition (MET), into the functional units of the kidney, the nephrons. MET is initiated by reciprocal induction between the ureteric bud (UB) and the blastema, followed by polarisation and differentiation of the blastema via gradually maturing epithelial structures, such as renal vesicles, comma‐shaped bodies (CSB) and S‐shaped bodies (SSB), that ultimately form tubules and glomeruli 1. Wilms tumours result from a disturbance of these processes with the accumulation of cells that retain features of the immature metanephric mesenchyme/blastema, with a variable degree of differentiation towards epithelial and stromal elements. This explains the tumour's classic triphasic histology. Tumours with a predominance of blastemal cells after treatment are associated with inferior prognosis, as are tumours with regions harbouring pleomorphic and disorganised cells, a feature referred to as diffuse anaplasia 2. Wilms tumour patients currently have an overall survival rate approaching 90%, but this comes at the price of heavy chemotherapy, often resulting in severe and/or life‐long side effects 3, 4. Identification of agents that could induce differentiation of Wilms tumour cells into nephrogenic structures could be a future route towards reducing cytotoxic treatment.
Somatic mutations and chromosomal rearrangements have been associated with high‐risk histological features and inferior outcome in Wilms tumour patients. Among those are hemizygous deletions in chromosome arm 16q 5. The smallest region of genomic overlap between these deletions has been shown to harbour the Iroquois homebox B (IRXB) gene cluster, consisting of IRX3, IRX5 and IRX6 6. IRX3 and IRX5 encode proteins that have recently been shown to be essential for several aspects of embryogenesis, such as the development of the nervous system, heart and skeleton 7, 8, 9, 10, 11. The Iroquois genes Irx3 and Irx5 are both expressed in the developing mouse kidney 12, but so far, no specific function for Irx5 in nephrogenesis has been reported 13. Irx3, on the other hand, has been shown to be important in nephron formation and segmentation in both Xenopus and zebrafish 14, 15, 16. This background encouraged us to further study the roles of IRX3 and IRX5 in Wilms tumour and kidney development with the ultimate purpose of identifying signalling pathways that could be manipulated for the induction of Wilms tumour differentiation.
Materials and methods
Mouse models, patient material and human cell lines
One wild type (wt), four Irx3/Irx5 +/− and four Irx3/Irx5 −/− E13.5 foetal mouse kidneys were available for analysis 11. Mouse experiments were performed following ethics permit ‘Mouse Limb Development’ #33868 (Mouse) approval by the Hospital for Sick Children Animal Care Committee, Toronto, Canada. Use of patient material was approved by the Regional Ethics Review Board with approval reference numbers L605‐2005 (biobanking), L289‐2011 (molecular analyses of tumours) and L796‐2017 (analysis of anonymised post‐mortem material), according to the Helsinki Declaration of 1975, as revised in 1983. Material from 39 primary Wilms tumours, two anonymised postnatal kidneys and five anonymised foetal kidneys were analysed (see supplementary material, Table S1). The Wilms tumour WiT49 cell line, kindly provided by Dr. Yeger at the Laboratory of Medicine and Pathobiology, University of Toronto, Canada 17, has been repeatedly characterised by our lab 18, 19. WiT49 cells were used to create IRX3 and IRX5 knockout cells through targeted genome editing to create IRX3 −/− and IRX5 −/− cells as detailed in the supplementary material, Supplementary materials and methods and Table S2. In vitro, cells were cultured in DMEM F:12 1:1 supplemented with 10% foetal bovine serum and 1% penicillin–streptomycin.
Orthotopic xenograft transplantation and magnetic resonance imaging
One million WiT49 wild‐type, IRX3 −/− or IRX5 −/− cells in 20 μl PBS were injected into the left kidney of female NSG mice (4–6 weeks old; Charles River Laboratories, Wilmington, MA, USA) as described previously 19. Thirteen mice were injected with WiT49 wild‐type cells, 14 with IRX3 −/− cells and 9 with IRX5 −/− cells (see supplementary material, Table S3). Magnetic resonance imaging (MRI) was performed 12 weeks after tumour cell injection as described previously (9.4 T horizontal bore, Lund University Bioimaging Center, Sweden; 20). MRIs were scrutinised using the Sante DICOM viewer 3D (Santesoft, Nicosia, Cyprus). Tumour volumes were calculated using the DICOM viewer software OsiriX (Pixmeo SARL, Switzerland). Volume was registered as 0 if no tumour was visible or if rendered immeasurable by being present in one MRI section only (see supplementary material, Table S3). Mice were sacrificed 3 months after xenotransplantation. Xenograft animal studies were performed according to ethics permit M11‐15 (Malmö‐Lund Animal Research Ethics Committee, Lund, Sweden).
Immunohistochemistry and histological imaging
Paraffin‐embedded tissue was deparaffinised and prepared for immunohistochemistry (IHC) according to standard procedures. Details about antibodies, their working solutions and procedures can be found in supplementary material, Table S4 and Supplementary materials and methods. Slides subjected to IHC or haematoxylin and eosin (H&E) staining were scanned using an Aperio ImageScope v12.1.0.5029 (Leica Biosystems, Wetzlar, Germany) or photographed with a digital SC50 camera (Olympus, Tokyo, Japan).
For murine embryonic kidneys, UB‐derived CALB1‐positive structures were distinguished from N‐cadherin‐positive CSB and SSB, while E‐cadherin was used to identify all epithelial foetal kidney components. CALB1‐immunostained sections were compared to consecutive sections stained for E‐cadherin to confirm the classification of CSB and SSB as opposed to UB‐derived structures. The total number (n) of CALB1‐ and N‐cadherin‐stained sections available for analysis in each group was: wild type: n = 3; Irx3/Irx5 +/−: n = 14; and Irx3/Irx5 −/−: n = 8. The Wilcoxon rank‐sum test was used for significance tests. For analysis of clinical Wilms tumour samples, a tissue microarray (TMA) with cores from 33 primary tumours was used for the evaluation of IRX3 and IRX5 protein expression 21. IRX3 and IRX5 were categorised as present or absent (see supplementary material, Table S1).
Xenograft Wilms tumours were evaluated for the percentage of tumour surface area consisting of neoplastic tubules. Positive WT1 expression identified tubular epithelial cells, whereas p53 positivity was used for demarcation of tumour borders in contrast to host mouse kidney tissue (see supplementary material, Figure S1). Thirteen wild‐type WiT49 tumours, 12 IRX3 −/− tumours [C2 (n = 6) and T5_40 (n = 6)] and 7 IRX5 −/− tumours [T5_34 (n = 2), T5_71 (n = 3) and T5_77 (n = 2)] provided sufficient tumour material for analysis (see supplementary material, Table S3). Evaluation of WNT5A expression was performed on sections from four WiT49 wild‐type, six IRX3 −/− and two IRX5 −/− xenograft tumours (see supplementary material, Table S3). All tumour areas containing stroma were digitalised, and all stroma cells [wild type (n = 3054 cells), IRX3 −/− (n = 5010 cells) and IRX5 −/− (n = 1115 cells)] were scored as positive or negative for WTN5A protein expression. Fisher's exact test (two‐sided) was used to evaluate any differences between groups.
RNA and protein extraction and gene and protein expression analyses
WiT49 wild‐type (n = 5), IRX3 knockout [C2 (n = 3) and T5_40 (n = 3)] and IRX5 knockout (one each of T5_34, T5_71, T5_77, T5_56 and T5_110) cells were seeded and grown for 48 h. Biological replicates were grown for WiT49, C2 and T5_40 in order to increase the number of samples for statistical analysis. For global gene expression analyses, whole transcriptome RNA sequencing was performed on an Ion Ampliseq (Thermo Fisher Scientific, Waltham, MA, USA) at the National Genomics Infrastructure, SciLife Uppsala Core Facility, Uppsala, Sweden. To explore active signalling pathways in IRX3 −/− and IRX5 −/− cells, gene expression data were analysed by Gene Set Enrichment Analysis (GSEA; 22, 23; GSEA 3.0 (http://www.broadinstitute.org/gsea/), Broad Institute, Cambridge, MA, USA). For targeted pathway analysis, Qlucore Omics Explorer 3.2 and 3.3 (Qlucore, Lund, Sweden) were used. Further details for gene expression analysis and information about protein extraction and Western blotting are provided in supplementary material, Supplementary materials and methods.
Cell proliferation assays
Cell proliferation assays were performed on the original WiT49 clone and its IRX3 −/− and IRX5 −/− clones using an MTS assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay, G3582, Promega, Madison, WI, USA) and IncuCyteZoom® (Essen BioScience, Ann Arbor, MI, USA). For details, see supplementary material, Supplementary materials and methods.
Results
Reduced nephron morphogenesis in Irx3/Irx5 knockout mice
To evaluate whether Irx3 and Irx5 influence normal mammalian kidney development, we first turned to an already established mouse model in which Irx3 and Irx5 had been either heterozygously or homozygously deleted 11. Given that Irx3 and Irx5 show redundant functions in mouse heart development, we chose to study double‐knockout embryos when probing for an effect on nephrogenesis 8, 24. Irx3/Irx5 −/− embryos die at E13.5 due to heart failure 8, 11. Therefore, we examined nephron formation during early kidney development (E13.5) and compared mice of wt or in which Irx3 and Irx5 had been either heterozygously or homozygously deleted (Figure 1). N‐cadherin immunostaining was used to identify cells and structures destined to become parts of proximal tubules, including SSB and CSB. Compared to wt and Irx3/5 +/− mice, maturing nephrogenic epithelial elements were close to absent in Irx3/Irx5 −/− mice (Figure 1M–R). CSB and SSB were significantly more prevalent in wt compared to Irx3/Irx5 +/− (p = 0.022) and Irx3/Irx5 −/− (p = 0.013) embryonal mouse kidneys (Wilcoxon rank‐sum test, Figure 1S). This role for Irx3/Irx5 in murine nephron formation encouraged further delineation of IRX3 and IRX5 in a context of human nephrogenesis and Wilms tumour.
Figure 1.
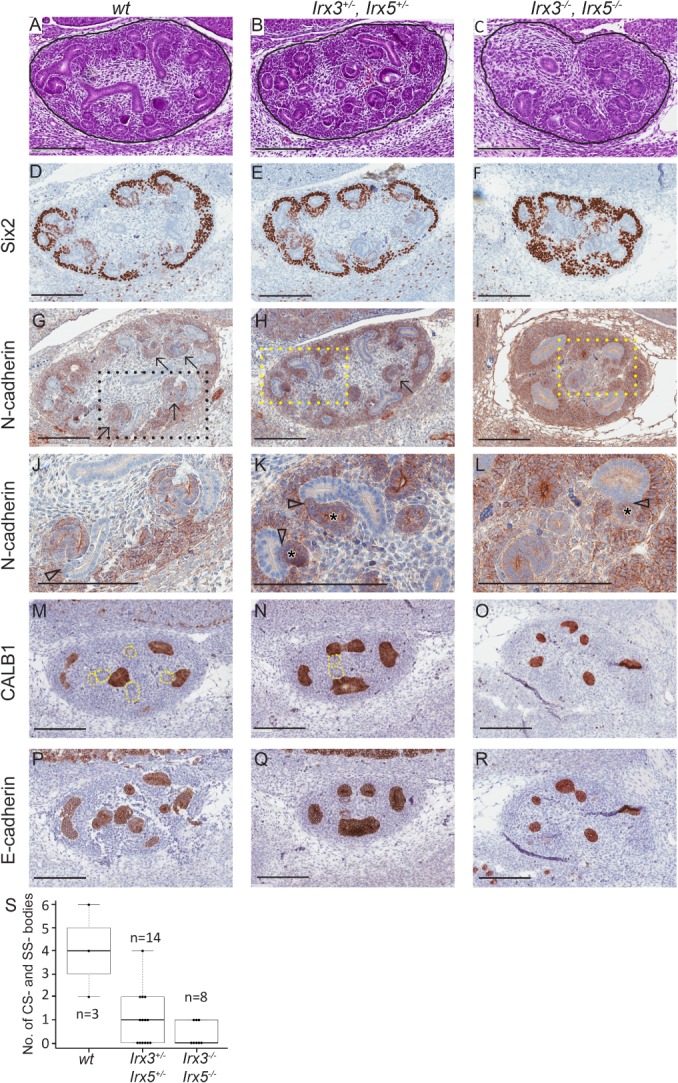
Reduced nephron morphogenesis in Irx3/Irx5 knockout mice. Representative images of Irx3 and Irx5 wt, heterozygous and homozygous knockout E13.5 foetal mouse kidney (FMK) sections stained with H&E (Htx; A–C). The CM/committed blastema, identified by Six2 expression was retained in all genotypes (D–F), although it was more extensively manifested in homozygous (Irx3 −/− Irx5 −/−) mice (F). Irx3/5 −/− mice, as opposed to wt and heterozygous mice, showed N‐cadherin positivity largely confined to cells of the CM as if differentiation through CSB and SSB had ceased (G–L). Specific labelling of UB structures with CALB1 (M–O), in combination with staining for all epithelial elements in the foetal kidney by E‐cadherin (P–R), confirmed that almost all epithelial structures were UB‐derived in Irx3/5 −/− mice (O and R). Irx3/Irx5 knockout mice maintained the ability to form some primitive nephrogenic tubules with a distinct lumen and could dock to UB‐derived structures, but the tails of the SSB were consistently missing in Irx3/Irx5 hetero‐ and homozygous knockout embryos, resulting in short tubules without curvature (K and L). Solid black lines demarcate the developing kidney from surrounding tissue (A–C). Arrows point to CSB and SSB in G and H. Content in rectangles with dashed lines in G–I are enlarged in J–L. Arrowheads point at docking sites between nephrogenic and UB‐derived epithelial structures, and asterisks (*) denote the lumen of nephrogenic tubules in K–L. Dotted lines in M and N correspond to E‐cadherin‐positive structures in P and Q. Scale bars correspond to 200 μm. All immunostains are brown. Box plot showing the median number and quartiles of CSB and SSB in FMK (S), where n denote the number of sections analysed for CSB and SCB in each group.
IRX3 and IRX5 are expressed in distinct tissue elements of foetal kidney and Wilms tumour
In human foetal kidney specimens (Figure 2), the nephrogenic epithelium was IRX3‐positive by IHC, whereas UB‐derived structures were negative (Figure 2A–C). Primitive nephrogenic structures of the nephrogenic zone, such as renal vesicles, CSB and SSB, as well as mature proximal tubules were IRX3‐positive (Figure 2D–F). The expression of IRX3 in cap mesenchyme (CM) was minimal (Figure 2D,E), and its expression increased with the time of gestation (Figure 2B,J,E), subsequently decreasing in the postnatal kidney (Figure 2F,L; see supplementary material, Figure S2A–D). IRX5 showed a similar expression pattern to IRX3 but was more prominently expressed in CM, renal vesicles and early‐stage nephrogenic tubules (Figure 2G,H and supplementary material, Figure S2E). IRX5 expression was retained in nephrons of postnatal normal kidney (Figure 2I). IRX3 and IRX5 showed predominantly cytoplasmic expression, with little nuclear expression at early nephrogenesis and none in postnatal kidney (Figure 2J–M). Similar to that observed in the human foetal kidney, IHC analysis of a TMA containing cores from ≥30 primary Wilms tumours demonstrated that IRX3 was expressed in the epithelial elements of almost all tumours, compared to blastemal components in a minority of cases, and in stromal components only very rarely (Figure 2N–P, T and supplementary material, Figure S2F). IRX5 was also expressed in epithelial components, but contrary to IRX3, it was also commonly expressed in blastemal and stromal elements (Figure 2Q–T). The IRX3 and IRX5 proteins were detected in both nucleus and cytoplasm (Figure 2N–S). Taken together, these results indicated that IRX3 may play a role primarily during maturation of nephrogenic elements, while IRX5 activity is also present in the stromal and proliferative late blastemal/early epithelial cells in developing kidneys and Wilms tumours.
Figure 2.
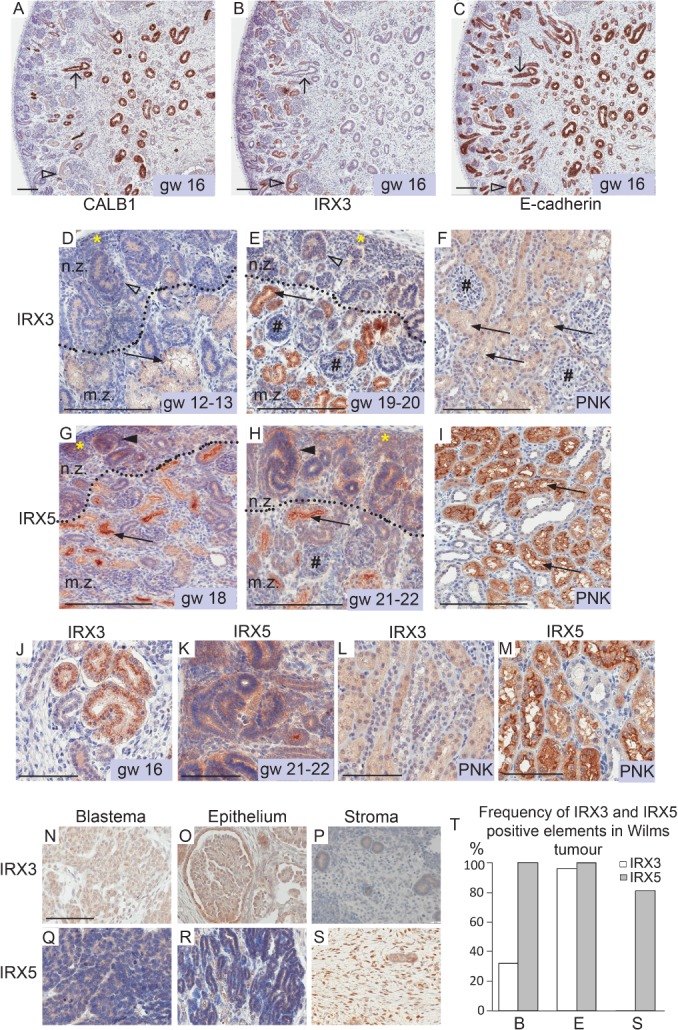
IRX3 and IRX5 are expressed in nephrogenic elements of human foetal kidney and distinct compartments of Wilms tumours. Expression of IRX3 and IRX5 proteins in human foetal and postnatal kidneys were identified by IHC. Consecutive sections of human kidney, at gestational week (gw) 16, were immunostained for CALB1 (A), IRX3 (B) and E‐cadherin (C). E‐cadherin was used for the detection of all epithelial structures of the kidney, and UB‐derived structures were characterised by CALB1 expression. Arrows with open heads point at CALB1‐positive/UB‐derived epithelial elements negative for IRX3. Open arrowheads point at IRX3 and E‐cadherin‐positive nephrogenic structures, which are negative for CALB1. IRX3‐ (D, E) and IRX5 (G, H)‐positive elements in human foetal (D, E and G, H) and postnatal kidney (PNK; F, I). Magnification of previous figures to illustrate cytoplasmic staining of IRX3 and IRX5, where scale bars correspond to 100 μm (J–M). Dotted lines indicate a border between the immature subcapsular nephrogenic zone (n.z.) and the more mature central zone (m.z.) of the developing kidney. Yellow asterisks denote cap/metanephric mesenchyme. Arrows point at the ruffled lumen of proximal tubules. Open arrowheads point at IRX3‐positive CSB and SSB. Filled arrowheads in G–H denote IRX5‐positive CSB and SSB. IRX3‐ (E, F) and IRX5 (H)‐negative glomeruli are denoted by hash (#). Scale bars correspond to 200 μm. N–S: IRX3 and IRX5 protein expression evaluated by IHC in Wilms tumour (WT) histological sub‐compartments from which representative areas are shown (N–P and Q–S, respectively). Scale bars correspond to 100 μm. All immunostains are brown. (T) Bar chart summarising the frequency of IRX3‐ and IRX5‐positive blastemal (B), epithelial (E) and stromal (S) elements in primary Wilms tumours based on examination of a WT TMA (IRX3, n = 33 primary WTs; IRX5, n = 30 primary WTs; Table S1).
IRX3 and IRX5 knockout Wilms tumour cells have contrasting phenotypes in vitro
To elucidate the functionality of IRX3 and IRX5 in Wilms tumour development, targeted genome editing of IRX3 and IRX5 was performed on the human WiT49 Wilms tumour cell line, chosen as a model system as it retains the multi‐phasic histology of Wilms tumours when orthotopically xenografted into mice 17, 19, 25, 26. Most importantly, the model enables the evaluation of tumour tubule formation, corresponding to Wilms tumour epithelial differentiation. Two IRX3 and five IRX5 knockout clones were successfully created (see supplementary material, Figure S3, Table S2). Attempts were made for the generation of IRX3/IRX5 double‐knockout clones, but these were unsuccessful. In vitro, there were clear morphological differences between the knockout cell lines (see supplementary material, Figure S4A–F), with IRX5 −/− cells growing in epithelial monolayers, while IRX3 −/− and wt cells had a fibroblast‐like appearance and formed tumour spherules. IRX5 −/− knockout clones showed decreased proliferation by two independent methods compared to WiT49 and IRX3 −/− cells (see supplementary material, Figure S5). RNA sequencing of cultured cells (see supplementary material, Table S5), followed by GSEA, demonstrated signatures of cell cycle regulators and progenitor cell renewal in IRX3 −/− cells (see supplementary material, Table S6A and Figure S4G). In contrast, the expression of genes involved in nephrogenic maturation, such as cell polarity, epithelial sheet formation and development, was significantly enriched for in IRX5 −/− cells (see supplementary material, Figure S4H–J and Table S6B,C).
Lack of IRX3 blocks tubular maturation and promotes tumourigenesis in vivo
To simulate Wilms tumourigenesis in vivo, we performed orthotopic xenograft transplantation of 1 × 106 wt, IRX3 −/− or IRX5 −/− WiT49 cells into the left kidney of immunodeficient NSG (NOD scid gamma) mice. MRI 12 weeks after transplantation repeatedly showed bulky tumour development from wt and IRX3 −/− xenograft cells, with no difference in tumour size between the genotypes (Figures 3A,B and 4A; p = 0.56, Wilcoxon rank‐sum test). The tumours were readily visible macroscopically when the mice were sacrificed at 14–15 weeks after transplantation (Figure 3D,E,G,H). Histologically, both wt and IRX3 −/− xenograft tumours were dominated by a large mesenchymal/stromal compartment (Figures 3J,K and 4C–H), readily discernible from the murine host kidney cells through the endogenous accumulation of mutated p53 in WiT49 cells 17. Wt and IRX3 −/− tumours grew mostly in a sarcomatous fashion, displacing or engulfing nephrons and collecting ducts of the host kidney (Figures 3M,N and 4D,G). The areas of tumour tubules in IRX3 −/− tumours were significantly reduced compared to those in wt tumours in accordance with a role for IRX3 in tubular maturation (Figure 4B,E,H; p = 0.0002, Wilcoxon rank‐sum test).
Figure 3.
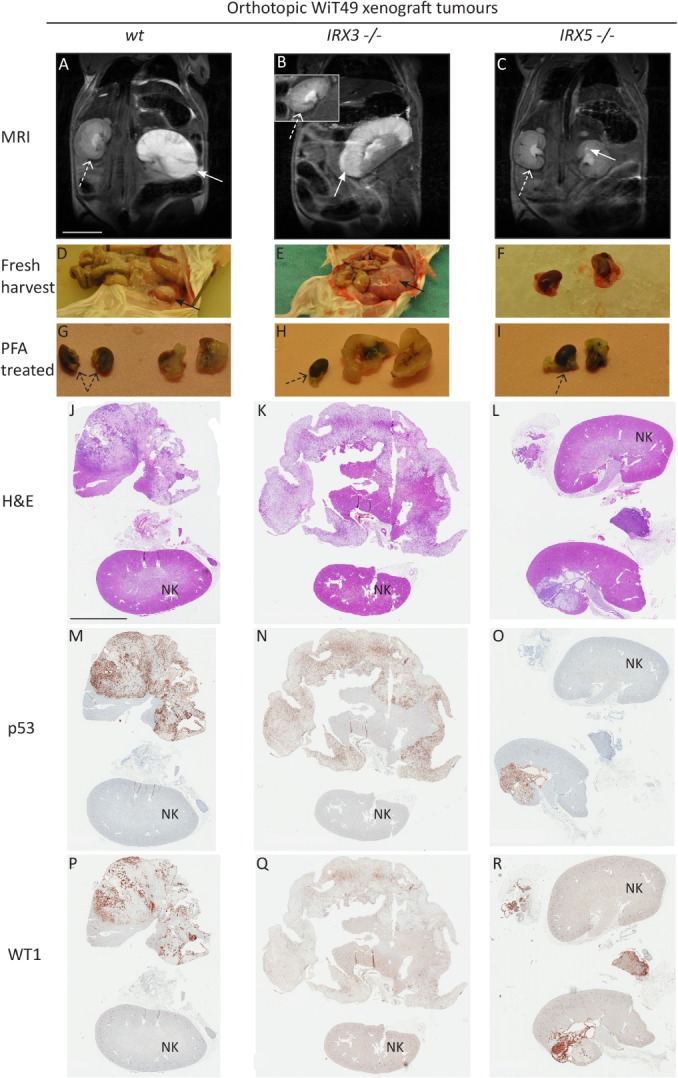
Distinct roles of IRX3 and IRX5 in orthotopic Wilms tumour xenografts. NSG mice were monitored by MRI 12 weeks after xenotransplantation of wt (A), IRX3 −/− (B) or IRX5 −/− (C) human Wilms tumour WiT49 cells into the left kidney. Arrows point at the tumour‐afflicted left kidney (A, B, C). Dashed arrows point at the unaffected right kidney (inset in B). Tumours appear as white areas with high signal intensity. Scale bar corresponds to 1 cm. Xenograft tumours were harvested and documented approximately 14 weeks after transplantation (D–F), followed by fixation of both kidneys in paraformaldehyde (PFA, G–I). Asterisks (*) denote visceral fat of the left kidney (F, I). Black dashed arrows point at unaffected right kidneys. Visualisation of tumour histology was carried out by H&E stains of xenograft tumours (J–L). IHC p53 positivity demonstrates tumour areas (M–O), whereas WT1 nuclear positivity enhances epithelial tumour elements (P–R). Scale bar corresponds to 5 mm. NK, normal kidney. All immunostains are brown.
Figure 4.
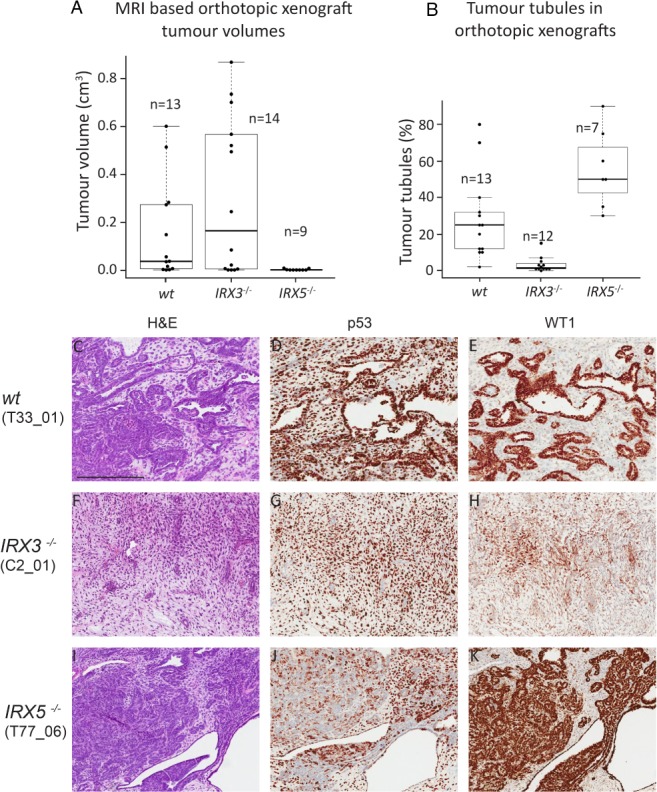
Loss of IRX3 promotes proliferative mesenchymal xenograft tumours, whereas loss of IRX5 stimulates differentiation of tumour tubules. (A) Tumour volumes based on MRI of NSG mice 12 weeks after xenograft transplantation with wt, IRX3 and IRX5 knockout orthotopic Wilms tumour xenografts. (B) The area of each tumour consisting of neoplastic tubules in wt, IRX3 and IRX5 knock out orthotopic xenografts. Boxplot represents median values and quartiles. (C–K): Detailed histological view of Wilms tumour orthotopic xenograft tumours with different IRX status, visualised by H&E staining (C, F, I) and IHC staining for p53 (D, G, J) and WT1 (E, H, K). The epithelial compartment of xenograft tumours was visualised by nuclear WT1 positivity, whereas p53 demarcated tumour cells from mouse host tissue. Within parentheses ( ) are the denotations of the specific xenograft tumours, listed in supplementary material, Table S3. Scale bar corresponds to 400 μm. All immunostains are brown.
Loss of IRX5 stimulates and maintains a differentiated state
Tumours emerging from grafted IRX5 −/− cells were either very small or were undetectable by MRI (see supplementary material, Table S3). They were rarely visible macroscopically upon harvest (Figure 3C,F,I) and their volumes were significantly smaller than wt and IRX3 −/− tumours as measured by MRI (Figure 4A; p = 0.002 and p = 0.009, respectively, Wilcoxon rank‐sum test) and microscopic analysis, respectively (Figure 3L,O,R compared to Figure 3J,K,M,N,P,Q). In fact, some IRX5 −/− tumours were only possible to ascertain after p53 and WT1 IHC (see supplementary material, Figure S6A–F). The size of the tumours indicated a low proliferation rate of IRX5 −/− cells, which recapitulated the results shown in vitro. Notably, clone T5‐77 also formed small tumours despite its lack of suppressed proliferation in vitro (Figure 4A and see supplementary material, Figure S5). IRX5 −/− tumours were histologically dominated by the epithelium (Figures 3R and 4K and see supplementary material, Figure S6C,F), with significantly more tumour tubules compared to both wt and IRX3 −/− tumours (p = 0.02 and p = 0.0004, Wilcoxon rank‐sum test; Figure 4B–K). Their mesenchymal/stromal compartments were minimal or absent (see supplementary material, Figure S6A–F). The two proteins did not appear to compensate for each other's expression in the xenograft knockout tumours, suggesting that other pathways downstream of the IRX proteins mediate the contrasting phenotypes (see supplementary material, Figure S6G–J).
IRX3 and IRX5 knockout affects WNT and Hippo signalling in different ways
To unravel the signalling pathways involved in the distinct phenotypes of IRX3 −/− and IRX5 −/− tumours, targeted gene expression analyses of key genes for nephrogenesis was performed 27. Here, IRX5 −/− cells had a signature more similar to that of wt WiT49 cells than to IRX3 −/− cells (Figure 5A). This was consistent with the higher prevalence of nephrogenic tubules in wt and IRX5 −/− tumours compared to that of IRX3 −/− tumours. Both WNT and Hippo signalling are known to be involved in nephrogenesis through, for example, planar cell polarity (PCP), which is important for tubule formation 28. Gene sets involved in both these pathways exhibited distinct expression signatures at comparison between IRX3 −/− and IRX5 −/− cells [Figure 5B,E; multi‐group comparison by analysis of variance (ANOVA)]. WNT5A is a key player in kidney development. Besides acting via the non‐canonical PCP WNT pathway 29, it can signal through the WNT receptor Fzd3 (reviewed in 30, 31, 32). Downstream of WNT receptor Fzd3, the cell adhesion molecule Alcam acts via the non‐canonical WNT/JNK pathway to affect proximal tubule development 33. Strikingly, WNT5A, FZD3 and ALCAM were all upregulated in IRX5 −/− cells (Figure 5B,C). Expression data suggested that canonical β‐catenin WNT signalling was suppressed in IRX5 −/− cells as genes involved in inhibition of β‐catenin (CTNNBIP1, DKK3 and KREMEN1) were upregulated in IRX5 −/− cells, and CTNNB1 itself was downregulated (Figure 5B). The reverse was true for IRX3 −/− cells (Figure 5B).
Figure 5.
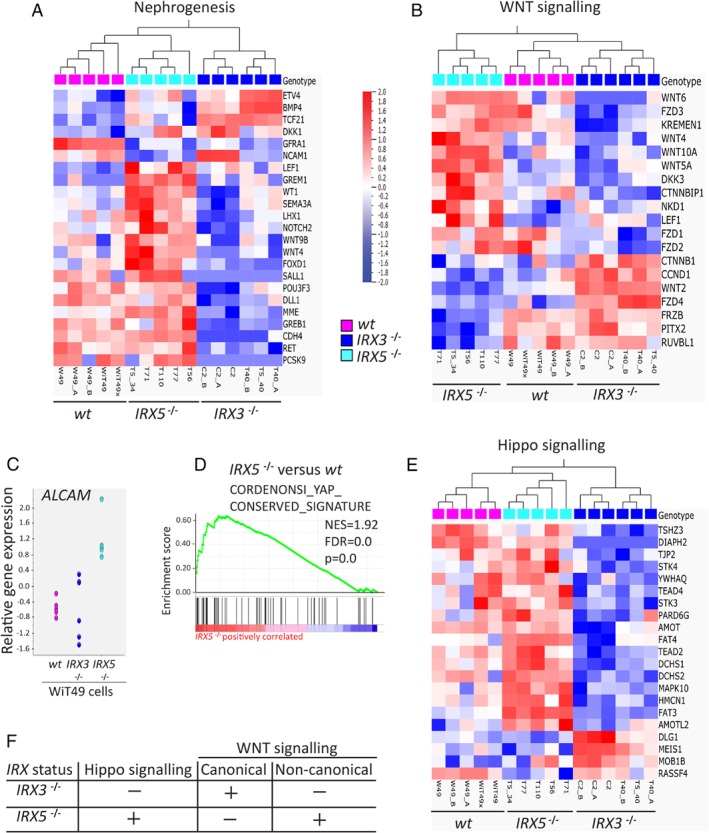
WNT and Hippo signalling‐related genes are differently expressed between IRX3‐ and IRX5‐ko WiT49 cells. (A) Nephrogenesis‐related genes (23/54 genes, p = 0.02) and (B) WNT signalling‐related genes (19/84 genes, p = 0.01) were differently expressed between wt, IRX3 −/− and IRX5 −/− WiT49 cells at multi‐group comparison by ANOVA (false detection rate = 0.05). Extracted genes were subjected to hierarchical clustering, and results were graphically displayed as heat maps. For further details, see Materials and Methods and supplementary material, Supplementary materials and methods and Table S7. Scatter plot displaying gene expression of the key nephron tubulogenesis gene ALCAM (C). GSEA supported the role of Hippo signalling in IRX5 −/− WiT49 cells. (D; GSEA run against the C6 oncogenic signatures gene sets). In addition, Hippo‐related genes (21/76 genes, p = 0.01, multi‐group comparison by ANOVA) were differently expressed between wt, IRX3 −/− and IRX5 −/− WiT49 cells (E). The gene expression differences between IRX3 −/− and IRX5 −/− cells imply that Hippo and non‐canonical WNT signalling are promoted in IRX5 −/− cells, whereas in IRX3 −/− cells, canonical WNT signalling is activated, and Hippo signalling is inhibited (F). The heat map scale units apply to all heat maps (A, B and E).
Hippo signalling is involved in the regulation of organ size, and it consists of a cascade of negative growth regulators. Hence, active Hippo signalling is considered important for nephrogenesis and the suppression of tumour formation 34. The expression of genes involved in Hippo signalling was significantly enriched in IRX5 −/− cells (Figure 5D and see supplementary material, Table S6D). In addition, specific genes important for the activation of Hippo signalling, such as AMOT, AMOTL2, DCHS1, DCHS2 and the PCP‐related FAT3 and FAT4, were upregulated in IRX5 −/− cells compared to IRX3 −/− cells (Figure 5E). Taken together, our data suggested that canonical WNT signalling was promoted in IRX3 −/− cells, whereas Hippo and non‐canonical WNT signalling were inhibited. The opposite was true for IRX5 −/− cells (Figure 5F).
WNT5A is a likely effector of nephrogenic tubule maturation
WNT5A is a key effector molecule in nephrogenesis and is also integrated with Hippo signalling 29, 35. It also plays a role in tumorigenesis and has been used to enforce the differentiation of colon cancer cells 36. WNT5A mRNA expression was higher in IRX5 −/− compared to IRX3 −/− cells (Figure 5B), while WNT5A protein was expressed in all histological elements of WiT49‐derived tumours irrespective of IRX3/5 status (Figure 6A–F). WNT5A‐positive mesenchymal cells were significantly more common in wt and IRX5 −/− as opposed to the poorly differentiated IRX3 −/− tumours (Fisher's exact test (two‐sided), p < 0.01, supplementary material, Figure S7), in line with its lower mRNA expression in the latter. WNT5A was primarily expressed in the maturing epithelial structures of the human foetal kidney (Figure 6G). In Wilms tumours, WNT5A showed the most prominent protein expression in tumour tubular epithelium, along with areas of surrounding stromal cells (Figure 6H and supplementary material, Figure S8A–F). Blastema was repeatedly negative (n = 6; Figure 6H). Taken together, these data implicate a collaborative role of WNT5A and IRX3 in the promotion of tumour tubulogenesis.
Figure 6.
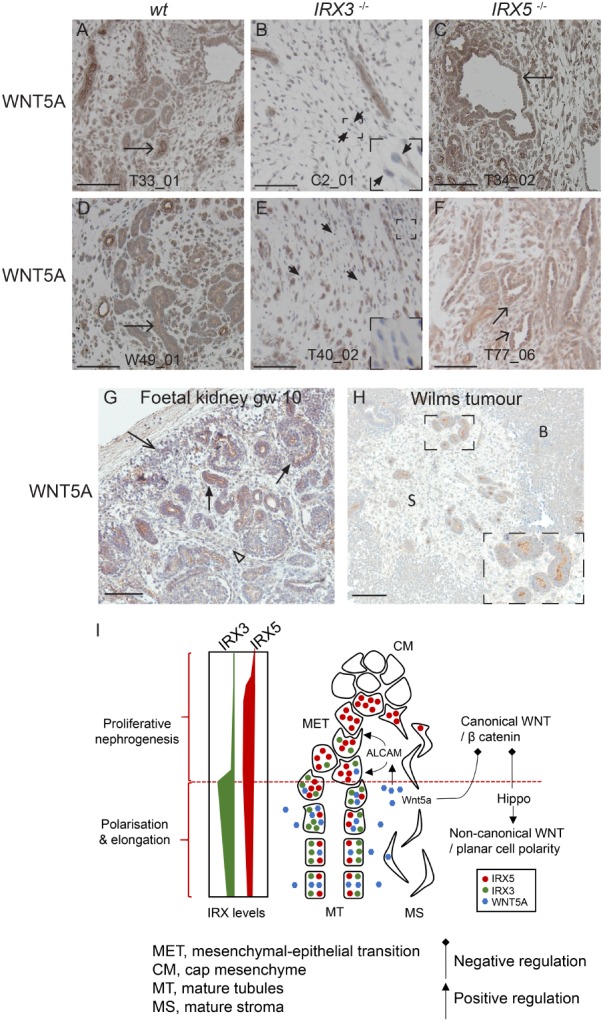
WNT5A is a likely differentiation‐promoting agent in Wilms tumour. WNT5A detected by IHC in WiT49‐based IRX3 and IRX5 knockout xenografts (A–F). Arrows point at WNT5A‐positive tumour tubules. Short arrows point at WNT5A‐negative stroma cells. Areas magnified ×3 of WNT5A‐negative stroma cells are inserted in (B) and (E). WNT5A expression in human foetal kidney at gestational week (gw) 10 and Wilms tumour (G and H, respectively). Arrowhead points at WNT5A‐positive stroma. Arrow with open arrowhead points at WNT5A‐positive metanephric mesenchyme. Arrows with filled arrowheads denote primitive nephrogenic elements. The insertion in H is a ×2 magnification of tumour tubules (H). Scale bars correspond to 100 μm. S = stroma. B = blastema. All immunostains are brown. (I) Cartoon depicting the proposed interplay between IRX3, IRX5 and WNT5A in nephrogenesis. IRX5 acts together with canonical/WNT‐β catenin signalling and disturbed Hippo signalling to maintain proliferative nephrogenesis. Polarisation and elongation are obtained via the integration of active Hippo and non‐canonical WNT signalling, where IRX3 and WNT5A, via stimulation of ALCAM, are crucial. Disturbed IRX3 and IRX5 expression, epitomised by Wilms tumour, inhibits MET and polarisation of immature mesenchyme. According to our model, exogenous WNT5A could potentially override the differentiation block present in Wilms tumour.
Discussion
Our study demonstrates that Irx3 and Irx5 are both expressed in primitive nephrons and regulate essential components of kidney formation in mice and humans. The expression pattern of IRX3 suggests its involvement primarily in the maturation of human nephrogenic structures, in line with the role of the Iroquois transcription factors in the patterning of nephrogenic tubules in the pronephros of zebrafish and amphibians 14, 15, 16. In contrast, the IRX5 expression pattern indicated a role for it primarily in the maintenance of a proliferative pool of nephrogenic cells and the cells of early tubule formation. Apart from our study, the evidence for a role of Irx5 in nephrogenesis is sparse 12. In accordance with the distinct roles of the two proteins in normal nephrogenesis, the IRX3 −/− and IRX5 −/− tumour cell phenotypes were in stark contrast to each other. IRX3 −/− tumours had a shortfall of nephrogenic organisation with an extensive mesenchymal stromal element, whereas IRX5 −/− tumours showed a differentiated nephrogenic pattern. Thus, our data strongly pointed to a differentiation‐promoting role for IRX3 in Wilms tumour and a role for IRX5 in maintaining a pool of cells with the proliferative features of the early metanephric mesenchyme in the WiT49 xenograft system. A similar role in primary Wilms tumour material would fit well with the fact that we detected IRX5 protein expression not only in tumour tubules but also in blastema and tumour stroma. However, the role of IRX3 in neoplasia is likely contextual as IRX3 rather blocks differentiation in acute leukaemia 37 and shows tumour‐promoting potential in hepatocellular carcinoma 38.
An earlier study on a small sample set of Wilms tumours indicated that low mRNA expression of both IRX3 and IRX5 correlates to poor prognosis in Wilms tumour 6. However, in the present study, loss of IRX5 resulted in small, well‐differentiated tumours, which suggests an oncogenic‐like role for IRX5 in Wilms tumour, well in accordance with a recently proposed role as an oncogenic driver in prostate and colorectal cancer 39, 40, 41. One plausible explanation for the seemingly contradictory role of IRX5 as a promotor of tumorigenesis on the one hand and the correlation of poor prognosis to its reduced mRNA expression on the other hand is its correlated expression with IRX3 in clinical material. In a situation where both genes are downregulated, it is feasible that the lack of differentiation caused by reduced IRX3 expression overrides the anti‐tumorigenic effects of reduced IRX5. What drives the expression of IRX5 in a Wilms tumour setting needs further investigation.
Our finding that IRX3 and IRX5 have central roles in nephrogenic differentiation highlights new possibilities to overcome the differentiation block that underlies Wilms tumour formation. The results presented here suggest that either the promotion of IRX3 signalling or inhibition of IRX5 signalling could be routes towards enforced differentiation in Wilms tumours. Disturbed Hippo signalling can cause nephron progenitors to differentiate into myofibroblasts 42, and stromal–epithelial communication can act via Hippo signalling components, such as Fat4, Fat3 and Dchs1/2, to regulate kidney progenitor cell differentiation 43, 44, 45. Thus, the bulky mesenchymal/stromal compartment of IRX3 −/− xenograft tumours could be partly due to a low Hippo signalling activity compared to IRX5 −/− tumours. A feasible way of targeting IRX3 or IRX5 is by indirect intervention with either WNT or Hippo signalling or both (reviewed in 28, 46).
WNT5A is involved in an alternative WNT pathway that is integrated with Hippo signalling 35. At the same time, it is known that WNT5A acts to suppress tumour formation in several cancer types 36, 47, 48. In colon cancer, it even induces differentiation of neoplastic cells 36. A previous study showed that WNT5A is downregulated in Wilms tumour 49. We specifically found lower WNT5A protein expression in stroma surrounding tumour tubules in IRX3 −/− and wild‐type WiT49 cells compared to IRX5 −/− cells, suggesting that it has a role in the induction of tubulogenesis in Wilms tumours. During organ development, Wnt5a is often secreted and directs PCP‐dependent cellular migration and patterning along concentration gradients 50. In our material, IRX3 and WNT5A both exhibited the strongest IHC staining levels in maturing epithelial elements. This implies a concentration gradient based on diffusion from epithelial structures into the surrounding stroma and CM. Diffused WNT5A could then promote epithelial differentiation via, for instance, ALCAM, in turn leading to secondary endogenous WNT5A production through autocrine‐positive feedback 33. An alternative explanation would be that WNT5A is negatively regulated by IRX5. However, this would be at odds with the coexistence of these two proteins in maturing renal tubules.
In summary, our data uniformly suggest a key role for IRX3 and IRX5 in mammalian nephrogenesis and Wilms tumour differentiation. We also find evidence for an important interplay between IRX3 and WNT5A in driving tubular maturation as summarised in Figure 6I. This renders WNT5A a potential treatment agent for the enforced differentiation of Wilms tumours.
Author contributions statement
LHM conceptualised the study, wrote the original draft and administered the project. LHM, JK, DB, JE, CJ, KL, SH, CH and SLM carried out and advanced methodology. LHM, SLM, JK and DG performed formal analysis. DG, LHM, JE, SLM, HY, KL and CJ carried out the investigation. DG,KL, SH, CH, DB and JE provided resources. DG, LHM and JK critically reviewed and edited the manuscript. LHM and DG supervised the study and acquired funding. All authors were involved in writing the paper and had final approval of the submitted and published versions.
SUPPLEMENTARY MATERIAL ONLINE.
Supplementary materials and methods
Supplementary figure legends
Figure S1. Haematoxylin eosin, p53, WT1 and nuclear morphology as guidelines for xenograft tumour demarcation and tubules
Figure S2. Extended IRX3 and IRX5 immunostaining
Figure S3. Identification of IRX3 and IRX5 ko WiT49 cell lines
Figure S4. IRX3 and IRX5 ko Wilms tumour cells have contrasting phenotypes
Figure S5. Cell proliferation is reduced in IRX5 −/− cells
Figure S6. Minimal stromal and excessive epithelial elements in IRX5 −/− tumours
Figure S7. Different prevalence of WNT5A‐positive stroma cells in IRX3 −/− and IRX5 −/− tumours
Figure S8. Tumour tubules of Wilms tumours show WNT5A positivity
Table S1. Clinical Wilms tumour samples
Table S2. IRX3 and IRX5 WiT49 knock out clones
Table S3. Orthotopic xenograft tumours
Table S4. Immunohistochemistry reagent and antibody information
Table S5. Gene expression data
Table S6. Gene set enrichment analyses
Table S7. Nephrogenesis, WNT‐ and Hippo signalling related genes
Supporting information
Supplementary materials and methods
Supplementary figure legends
Figure S1. Haematoxylin and eosin, p53, WT1 and nuclear morphology as guidelines for xenograft tumour demarcation and tubules
Figure S2. Extended IRX3 and IRX5 immunostaining
Figure S3. Identification of IRX3 and IRX5 ko WiT49 cell lines
Figure S4. IRX3 and IRX5 ko Wilms tumour cells have contrasting phenotypes
Figure S5. Cell proliferation is reduced in IRX5 −/− cells
Figure S6. Minimal stromal and excessive epithelial elements in IRX5 −/− tumours
Figure S7. Different prevalence of WNT5A‐positive stroma cells in IRX3 −/− and IRX5 −/− tumours
Figure S8. Tumour tubules of Wilms tumours show WNT5A positivity
Table S1. Clinical Wilms tumour samples
Table S2. IRX3 and IRX5 WiT49 knockout clones
Table S3. Orthotopic xenograft tumours
Table S4. Immunohistochemistry reagent and antibody information
Table S5. Gene expression data
Table S6. Gene set enrichment analyses
Table S7. Nephrogenesis, WNT and Hippo signalling‐related genes
Acknowledgements
This study was supported by grants from the Swedish Research Foundation, the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Crafoord Foundation, the Royal Physiographic Society and the Medical Faculty of Lund University Sweden. The authors also acknowledge support from BioCARE, Sweden. Lund University Bioimaging Center (LBIC), Lund University and the Whole transcriptome RNA‐sequencing, National Genomics Infrastructure and SciLife Uppsala core facility, Sweden are gratefully acknowledged for providing experimental resources.
No conflicts of interest were declared.
References
*Cited only in supplementary material.
- 1. Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet 2007; 8: 791–802. [DOI] [PubMed] [Google Scholar]
- 2. Vujanic GM, Sandstedt B. The pathology of Wilms' tumour (nephroblastoma): the International Society of Paediatric Oncology approach. J Clin Pathol 2010; 63: 102–109. [DOI] [PubMed] [Google Scholar]
- 3. Kaste SC, Dome JS, Babyn PS, et al Wilms tumour: prognostic factors, staging, therapy and late effects. Pediatr Radiol 2008; 38: 2–17. [DOI] [PubMed] [Google Scholar]
- 4. Pritchard‐Jones K. Controversies and advances in the management of Wilms' tumour. Arch Dis Child 2002; 87: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grundy PE, Breslow NE, Li S, et al Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable‐histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol 2005; 23: 7312–7321. [DOI] [PubMed] [Google Scholar]
- 6. Mengelbier LH, Karlsson J, Lindgren D, et al Deletions of 16q in Wilms tumors localize to blastemal‐anaplastic cells and are associated with reduced expression of the IRXB renal tubulogenesis gene cluster. Am J Pathol 2010; 177: 2609–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bosse A, Zulch A, Becker MB, et al Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech Dev 1997; 69: 169–181. [DOI] [PubMed] [Google Scholar]
- 8. Gaborit N, Sakuma R, Wylie JN, et al Cooperative and antagonistic roles for Irx3 and Irx5 in cardiac morphogenesis and postnatal physiology. Development 2012; 139: 4007–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gomez‐Skarmeta JL, Modolell J. Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev 2002; 12: 403–408. [DOI] [PubMed] [Google Scholar]
- 10. Bonnard C, Strobl AC, Shboul M, et al Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nat Genet 2012; 44: 709–713. [DOI] [PubMed] [Google Scholar]
- 11. Li D, Sakuma R, Vakili NA, et al Formation of proximal and anterior limb skeleton requires early function of Irx3 and Irx5 and is negatively regulated by Shh signaling. Dev Cell 2014; 29: 233–240. [DOI] [PubMed] [Google Scholar]
- 12. Houweling AC, Dildrop R, Peters T, et al Gene and cluster‐specific expression of the Iroquois family members during mouse development. Mech Dev 2001; 107: 169–174. [DOI] [PubMed] [Google Scholar]
- 13. Marra AN, Wingert RA. Roles of Iroquois transcription factors in kidney development. Cell Dev Biol 2014; 3: 1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reggiani L, Raciti D, Airik R, et al The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev 2007; 21: 2358–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alarcon P, Rodriguez‐Seguel E, Fernandez‐Gonzalez A, et al A dual requirement for Iroquois genes during Xenopus kidney development. Development 2008; 135: 3197–3207. [DOI] [PubMed] [Google Scholar]
- 16. Wingert RA, Davidson AJ. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev Dyn 2011; 240: 2011–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alami J, Williams BR, Yeger H. Derivation and characterization of a Wilms' tumour cell line, WiT 49. Int J Cancer 2003; 107: 365–374. [DOI] [PubMed] [Google Scholar]
- 18. Gisselsson D, Lindgren D, Mengelbier LH, et al Genetic bottlenecks and the hazardous game of population reduction in cell line based research. Exp Cell Res 2010; 316: 3379–3386. [DOI] [PubMed] [Google Scholar]
- 19. Mengelbier LH, Bexell D, Sehic D, et al Orthotopic Wilms tumor xenografts derived from cell lines reflect limited aspects of tumor morphology and clinical characteristics. Pediatr Blood Cancer 2014; 61: 1949–1954. [DOI] [PubMed] [Google Scholar]
- 20. Braekeveldt N, Wigerup C, Gisselsson D, et al Neuroblastoma patient‐derived orthotopic xenografts retain metastatic patterns and geno‐ and phenotypes of patient tumours. Int J Cancer 2015; 136: E252–E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sehic D, Ciornei CD, Gisselsson D. Evaluation of CITED1, SIX1, and CD56 protein expression for identification of blastemal elements in Wilms tumor. Am J Clin Pathol 2014; 141: 828–833. [DOI] [PubMed] [Google Scholar]
- 22. Mootha VK, Lindgren CM, Eriksson KF, et al PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273. [DOI] [PubMed] [Google Scholar]
- 23. Subramanian A, Tamayo P, Mootha VK, et al Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang SS, Kim KH, Rosen A, et al Iroquois homeobox gene 3 establishes fast conduction in the cardiac His‐Purkinje network. Proc Natl Acad Sci U S A 2011; 108: 13576–13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bielen A, Box G, Perryman L, et al Dependence of Wilms tumor cells on signaling through insulin‐like growth factor 1 in an orthotopic xenograft model targetable by specific receptor inhibition. Proc Natl Acad Sci U S A 2012; 109: E1267–E1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li MH, Yamase H, Ferrer F. Characterization of a WiT49 cell line derived orthotopic model of Wilms tumor. Pediatr Blood Cancer 2010; 54: 316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlsson J, Holmquist Mengelbier L, Ciornei CD, et al Clear cell sarcoma of the kidney demonstrates an embryonic signature indicative of a primitive nephrogenic origin. Genes Chromosomes Cancer 2014; 53: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bernascone I, Martin‐Belmonte F. Crossroads of Wnt and Hippo in epithelial tissues. Trends Cell Biol 2013; 23: 380–389. [DOI] [PubMed] [Google Scholar]
- 29. Huang L, Xiao A, Choi SY, et al Wnt5a is necessary for normal kidney development in zebrafish and mice. Nephron Exp Nephrol 2014; 128: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dijksterhuis JP, Petersen J, Schulte G. WNT/frizzled signalling: receptor‐ligand selectivity with focus on FZD‐G protein signalling and its physiological relevance: IUPHAR review 3. Br J Pharmacol 2014; 17: 1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kikuchi A, Yamamoto H, Sato A. Selective activation mechanisms of Wnt signaling pathways. Trends Cell Biol 2009; 19: 119–129. [DOI] [PubMed] [Google Scholar]
- 32. Carroll TJ, Das A. Planar cell polarity in kidney development and disease. Organogenesis 2011; 7: 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cizelsky W, Tata A, Kuhl M, et al The Wnt/JNK signaling target gene alcam is required for embryonic kidney development. Development 2014; 141: 2064–2074. [DOI] [PubMed] [Google Scholar]
- 34. Wong JS, Meliambro K, Ray J, et al Hippo signaling in the kidney: the good and the bad. Am J Physiol Renal Physiol 2016; 311: F241–F248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park HW, Kim YC, Yu B, et al Alternative Wnt signaling activates YAP/TAZ. Cell 2015; 162: 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mehdawi LM, Prasad CP, Ehrnstrom R, et al Non‐canonical WNT5A signaling up‐regulates the expression of the tumor suppressor 15‐PGDH and induces differentiation of colon cancer cells. Mol Oncol 2016; 10: 1415–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Somerville TDD, Simeoni F, Chadwick JA, et al Derepression of the Iroquois homeodomain transcription factor gene IRX3 confers differentiation block in acute leukemia. Cell Rep 2018; 22: 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang P, Zhuang C, Huang D, et al Downregulation of miR‐377 contributes to IRX3 deregulation in hepatocellular carcinoma. Oncol Rep 2016; 36: 247–252. [DOI] [PubMed] [Google Scholar]
- 39. Myrthue A, Rademacher BL, Pittsenbarger J, et al The Iroquois homeobox gene 5 is regulated by 1,25‐dihydroxyvitamin D3 in human prostate cancer and regulates apoptosis and the cell cycle in LNCaP prostate cancer cells. Clin Cancer Res 2008; 14: 3562–3570. [DOI] [PubMed] [Google Scholar]
- 40. Liu T, Zhang X, Yang YM, et al Increased expression of the long noncoding RNA CRNDE‐h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther 2016; 9: 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mori K, Nakamura H, Kurooka H, et al Id2 determines intestinal identity through repression of the foregut transcription factor, Irx5. Mol Cell Biol 2018. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McNeill H, Reginensi A. Lats1/2 regulate yap/Taz to control nephron progenitor epithelialization and inhibit myofibroblast formation. J Am Soc Nephrol 2017; 28: 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Das A, Tanigawa S, Karner CM, et al Stromal‐epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol 2013; 15: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bagherie‐Lachidan M, Reginensi A, Pan Q, et al Stromal Fat4 acts non‐autonomously with Dchs1/2 to restrict the nephron progenitor pool. Development (Cambridge, England) 2015; 142: 2564–2573. [DOI] [PubMed] [Google Scholar]
- 45. Mao Y, Francis‐West P, Irvine KD. Fat4/Dchs1 signaling between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development 2015; 142: 2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McNeill H, Woodgett JR. When pathways collide: collaboration and connivance among signalling proteins in development. Nat Rev Mol Cell Biol 2010; 11: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Canesin G, Evans‐Axelsson S, Hellsten R, et al Treatment with the WNT5A‐mimicking peptide Foxy‐5 effectively reduces the metastatic spread of WNT5A‐low prostate cancer cells in an orthotopic mouse model. PLoS One 2017; 12: e0184418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prasad CP, Chaurasiya SK, Guilmain W, et al WNT5A signaling impairs breast cancer cell migration and invasion via mechanisms independent of the epithelial‐mesenchymal transition. J Exp Clin Cancer Res 2016; 35: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamimi Y, Ekuere U, Laughton N, et al WNT5A is regulated by PAX2 and may be involved in blastemal predominant Wilms tumorigenesis. Neoplasia 2008; 10: 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gao B, Song H, Bishop K, et al Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell 2011; 20: 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *. Gaj T, Guo J, Kato Y, et al Targeted gene knockout by direct delivery of zinc‐finger nuclease proteins. Nat Methods 2012; 9: 805–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *. Cong L, Ran FA, Cox D, et al Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Supplementary figure legends
Figure S1. Haematoxylin and eosin, p53, WT1 and nuclear morphology as guidelines for xenograft tumour demarcation and tubules
Figure S2. Extended IRX3 and IRX5 immunostaining
Figure S3. Identification of IRX3 and IRX5 ko WiT49 cell lines
Figure S4. IRX3 and IRX5 ko Wilms tumour cells have contrasting phenotypes
Figure S5. Cell proliferation is reduced in IRX5 −/− cells
Figure S6. Minimal stromal and excessive epithelial elements in IRX5 −/− tumours
Figure S7. Different prevalence of WNT5A‐positive stroma cells in IRX3 −/− and IRX5 −/− tumours
Figure S8. Tumour tubules of Wilms tumours show WNT5A positivity
Table S1. Clinical Wilms tumour samples
Table S2. IRX3 and IRX5 WiT49 knockout clones
Table S3. Orthotopic xenograft tumours
Table S4. Immunohistochemistry reagent and antibody information
Table S5. Gene expression data
Table S6. Gene set enrichment analyses
Table S7. Nephrogenesis, WNT and Hippo signalling‐related genes


