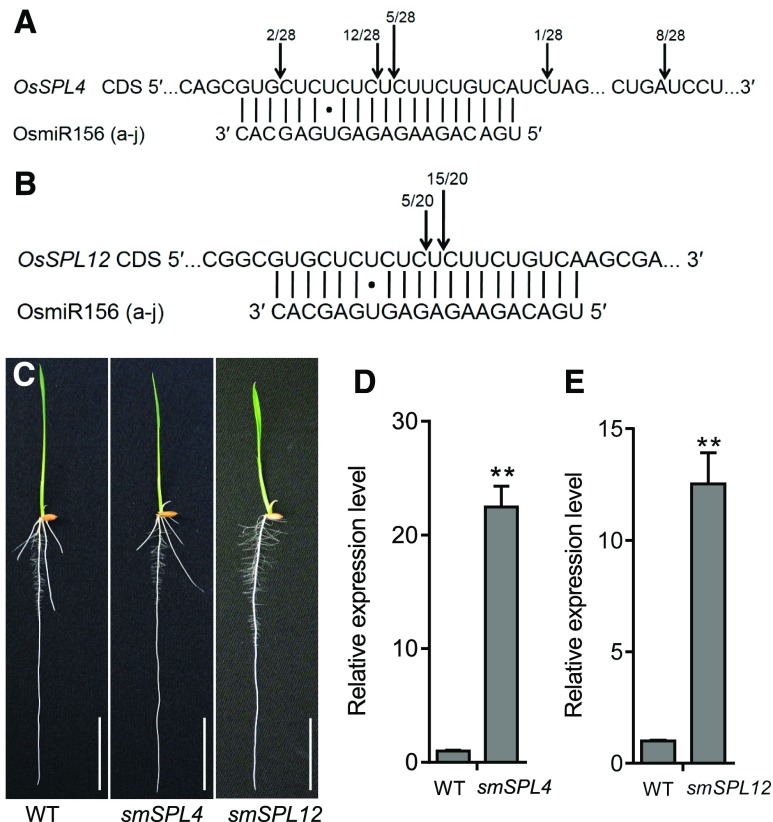
Figure 6.
OsmiR156-Regulated OsSPL12, but not OsSPL4, Inhibits Crown Root Development in Rice.
(A) and (B) OsmiR156-mediated cleavage sites in OsSPL4 (A) and OsSPL12 (B) mRNAs determined using RLM-RACE. The vertical lines represent the nucleotides that base pair with OsmiR156, and dots show the mismatched nucleotides. The positions corresponding to the 5′ ends of the cleaved OsSPL4 and OsSPL12 mRNAs determined using 5′ RACE are indicated by arrows, and the number of 5′ RACE clones corresponding to each site are shown.
(C) Phenotypes of 5-d-old ProSPL4:smSPL4 (smSPL4) and ProSPL12:smSPL12 (smSPL12) transgenic plants. Bars = 3 cm. WT, wild type.
(D) and (E) Quantitative real-time PCR analysis of the expression of OsSPL4 (D) and OsSPL12 (E) in 5-d-old transgenic plants. Data were normalized to those of the wild type ([WT]; the level of WT was set to 1). Values are means + SD (n = 3 independent pools of basal nodes, five plants per pool). **Significant difference compared with WT (P < 0.01; Student’s t test).