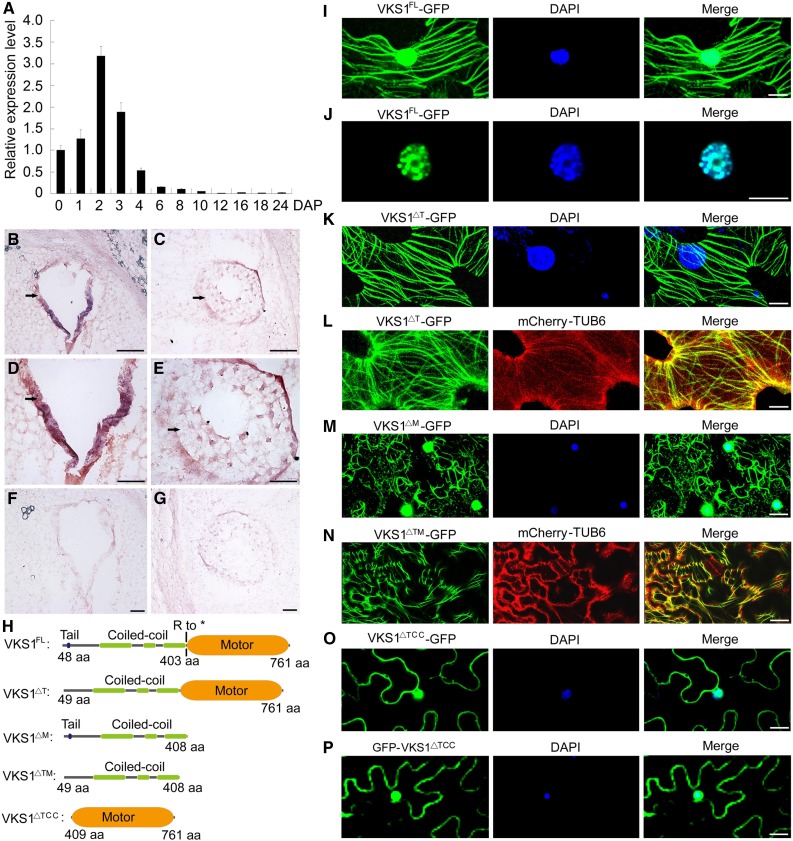
Figure 3.
The Gene Expression Pattern of VKS1 and Its Protein Subcellular Localization.
(A) RT-qPCR analysis of Vks1 during seed development. All expression levels are normalized to Actin. The expression level of Vks1 at 0 DAP is set to 1. Error bars represent sd from three biological replicates.
(B) to (G) RNA in situ hybridization of Vks1 in seeds at 2 and 3 DAP. Positive signals (shown in red) are clearly restricted to the coenocyte at 2 DAP ([B] and [D]) and endosperm cells engaging in cellularization at 3 DAP ([C] and [E]). When hybridized with sense probes, no signal is observed in the sections of seeds at 2 DAP (F) and 3 DAP (G). Bars = 50 μm.
(H) Schematic diagrams of constructs for subcellular localization. VKS1FL, VKS1△T, VKS1△M, VKS1△TM, and VKS1△TCC represent the full-length VKS1 protein and fragments lacking the tail, the motor, both the tail and motor domains, and both the tail and CC domains, respectively. aa, amino acids.
(I) Subcellular localization of VKS1FL-GFP. The nucleus is stained with DAPI.
(J) Localization of VKS1FL-GFP in the nucleus. The nucleus is stained with DAPI.
(K) Subcellular localization of VKS1△T-GFP. The nucleus is stained with DAPI.
(L) Colocalization of VKS1△T-GFP and mCherry-TUB6.
(M) Subcellular localization of VKS1△M-GFP. The nucleus is stained with DAPI.
(N) Colocalization of VKS1△TM-GFP and mCherry-TUB6.
(O) and (P) Subcellular localization of VKS1△TCC-GFP and GFP-VKS1△TCC. The nucleus is stained with DAPI.
For (I) to (P), bars = 10 μm.