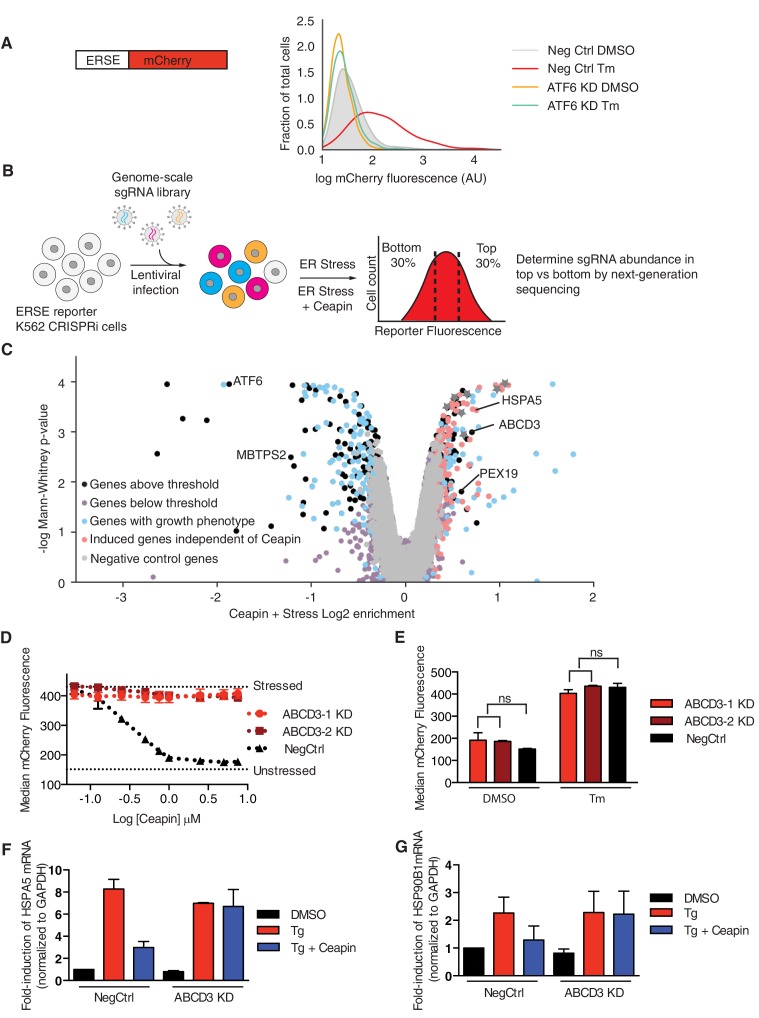
Figure 1. ABCD3 KD desensitizes cells to Ceapin-A7.
(A) Schematic of the ER stress element (ERSE) reporter cassette. K562 ERSE reporter cells were transduced with the indicated sgRNAs and treated with vehicle (DMSO) or tunicamycin (Tm) (6 μg/ml) for 16 hr. (B) Schematic of the CRISPRi screen to identify the target of Ceapin. K562 cells expressing the ERSE reporter were transduced with the sgRNA library. The population was then divided into two subpopulations, which were treated with Tm or Tm plus Ceapin-A7 at EC90 (3 μM) for 16 hr. Cells in the top and bottom thirds of mCherry fluorescence of each subpopulation (Tm-treatment and Tm + Ceapin-treatment) were collected by FACS and processed to measure the frequencies of sgRNAs contained within each. (C) Volcano plot of gene-reporter phenotypes and p values from CRISPRi screen. Negative control sgRNA targeted genes (gray), Ceapin-independent genes (red), genes with growth phenotypes (blue), and Ceapin hits (black) are indicated. (*) denotes chromatin architecture and remodeling related genes that impact reporter transcription. The reporter phenotypes and p values for genes in CRISPRi screen are listed in Figure 1—source data 1. (D) K562 ERSE reporter cells with individual ABCD3 sgRNAs or control sgRNA (NegCtrl) were treated with Tm and increasing concentrations of Ceapin-A7 for 16 hr. Reporter fluorescence was measured by flow cytometry and median values were plotted (N = 3, ± SD). (E) K562 ERSE reporter ABCD3 and NegCtrl KD cells were treated with DMSO or Tm and reporter activation was measured as in (D). (F and G) qPCR analysis of ATF6α target genes HSPA5 and HSP90B1, respectively. HepG2 CRISPRi NegCrl and ABCD3 KD cell lines were treated with DMSO, thapsigargin (Tg) (100 nM), and Tg with Ceapin (6 μM). Tg blocks the ER calcium pump and induces ER stress. Data plotted are mRNA levels for HSPA5 and HSP90B1 normalized to GAPDH and then compared to unstressed NegCtrl cells ± standard deviation of duplicate technical replicates of two biological replicates.
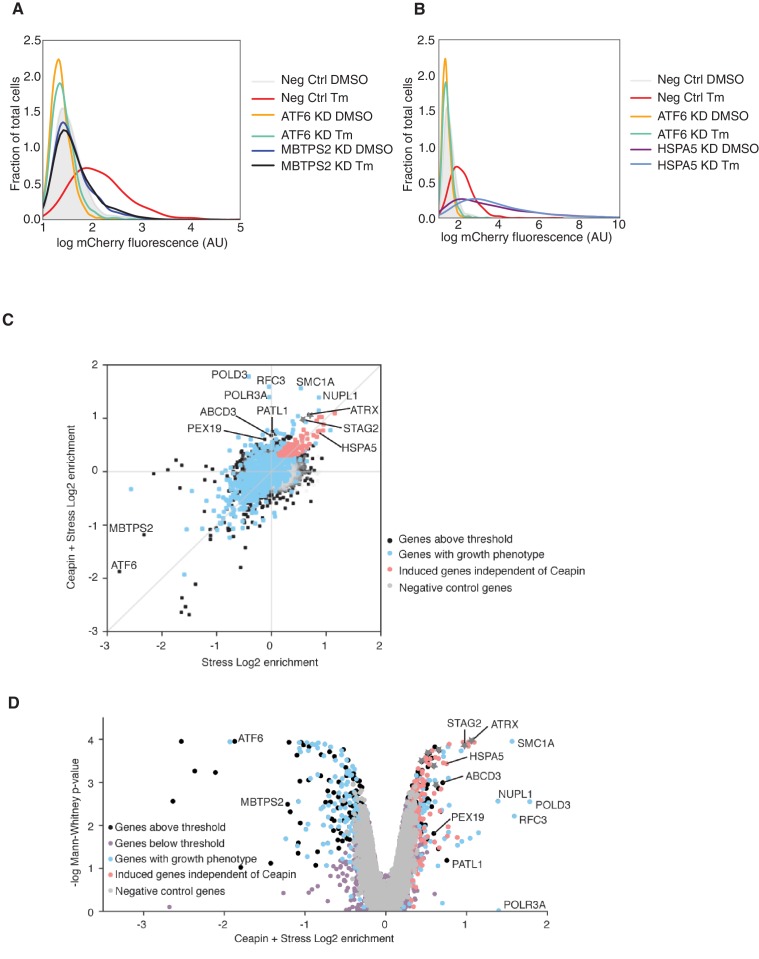
Figure 1—figure supplement 1. Genome-scale CRISPRi screen to identify molecular target of Ceapin.
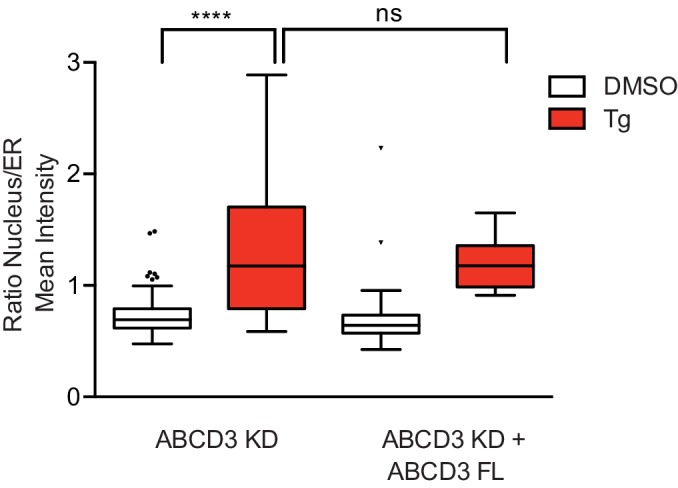
Figure 1—figure supplement 2. ABCD3 KD does not affect ATF6α nuclear translocation.