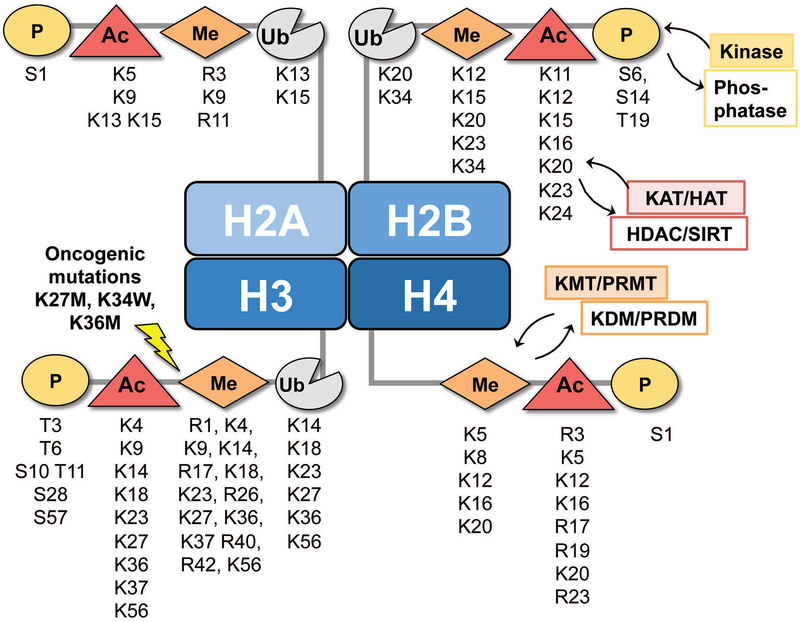
Figure 4: Post-translational modifications in histone proteins.
Post-translational modifications in nucleosomes are distributed over the N-terminal tails of the four core histone proteins H2A, H2B, H3 and H4. Each histone has multiple residues that can be covalently and reversibly modified by phosphorylation (P), acetylation (Ac), methylation (Me) or ubiquitination (Ub) using specific kinases, phosphatases, acetyl transferases (KAT/HATs), deacetylases (HDAC/SIRTs), histone methyl transferases (KMT/PRMTs) or demethylases (KDM/PRDMs). There are more than 300 distinct epigenetic regulators that generate, recognize or remove histone post-translational modifications.