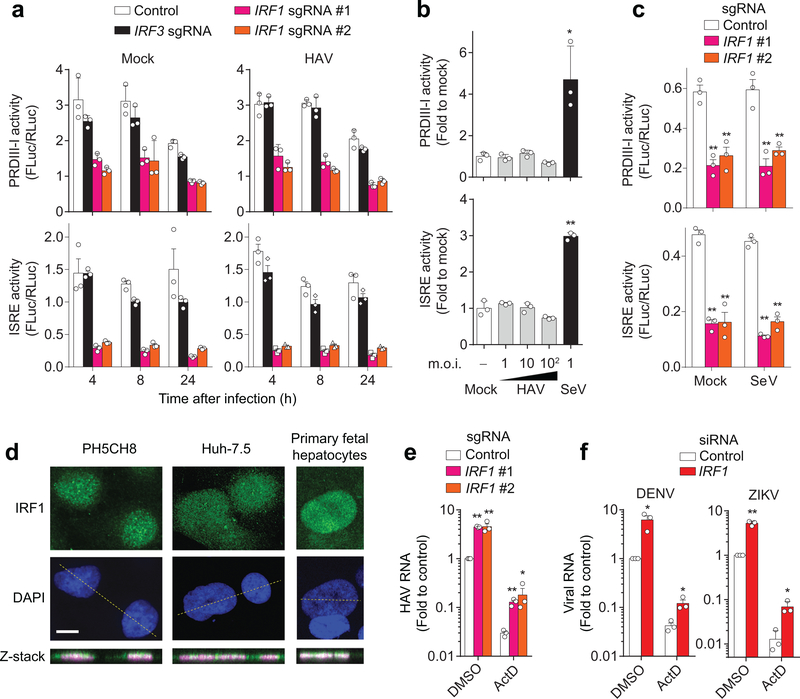
Figure 2. IRF1 constitutively activates basal transcription of PRDIII-I- and ISRE-dependent antiviral genes.
(a) Dual luciferase reporter analysis of 4×PRDIII-I-Luc (top panels) and ISRE-Luc (bottom panels) activities in mock- (left panels) and HAV-infected (right panels) PH5CH8 cells. Promoter activities in IRF1-sgRNA (#1, #2) vs. control or IRF3 sgRNA-expressing cells differed significantly (p < 0.01, two-way ANOVA with Dunnett’s multiple comparisons test). (b) Dose-response analysis of PRDIII-I (top) and ISRE (bottom) activities in HAV and SeV-infected, wildtype PH5CH8 cells at the indicated m.o.i. *p<0.05, **p<0.01 vs. mock (one-way ANOVA with Dunnett’s multiple comparisons test). (c) Dual luciferase reporter analysis of 4×PRDIII-I-Luc (top) and ISRE-Luc (bottom) activities in mock-infected Huh-7.5 cells. Note that SeV does not activate these promoters in Huh-7.5 cells. **p<0.01 vs. control (two-way ANOVA with Dunnett’s multiple comparisons test). (d) Nuclear localization of IRF1 in two different hepatic cell lines and primary human fetal hepatocytes. Data are representative of 2 independent experiments. Scale bar, 20 μm. (e) HAV RNA at 24 h p.i. in Huh-7.5 cells expressing IRF1 sgRNA pre-treated with actinomycin D (ActD, 5 μg ml−1) for 30 min before infection. **p<0.01, *p<0.05 vs. control (two-sided unpaired t-test). (f) DENV RNA at 18 h p.i. or ZIKV RNA at 24 h p.i. in IRF1-depleted Huh-7.5 cells pre-treated with actinomycin D (ActD, 5 μg ml−1) for 30 min before infection. *p<0.05, **p<0.01 vs. control (two-sided unpaired t-test). Data are means ± s.d. from 3 technical replicates representative of 2 independent experiments (a-d) or from 3 independent experiments (e, f). The precise p-values are shown in Supplementary Table 9.