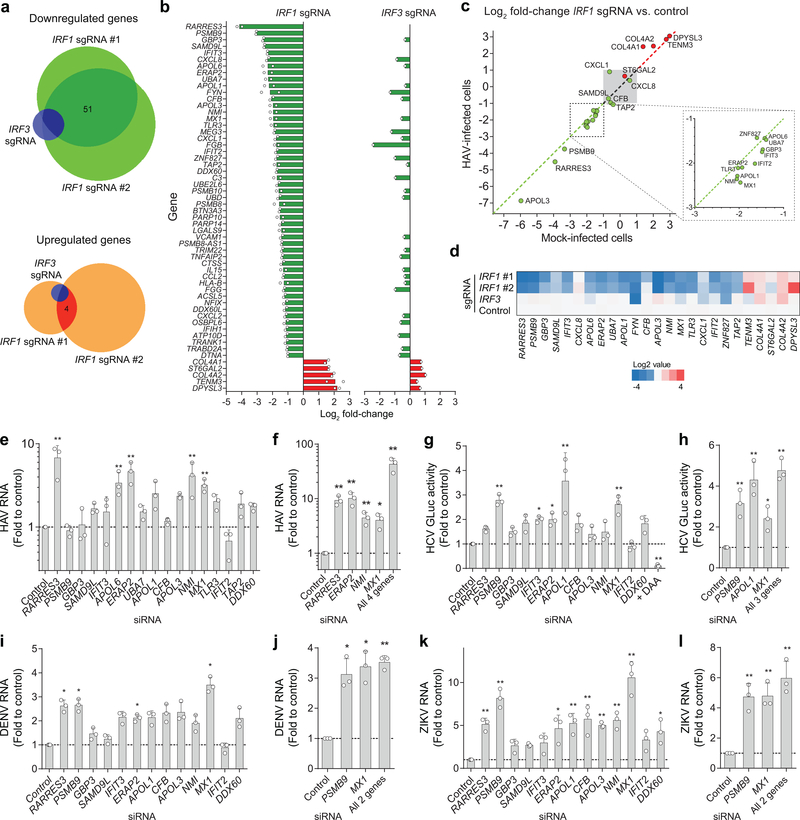
Figure 3. Shared and distinct antiviral activities of IRF1 effector genes identified by high-throughput RNA sequencing.
(a) The Venn diagram showing numbers of genes with expression changes of ≥ 2-fold for each knockout. (b) List of genes reduced >2-fold in IRF1 sgRNA-expressing cells, in comparison with IRF3 sgRNA-expressing cells. Values shown are means of fold changes of genes expressed in cells transduced with 2 independent IRF1 sgRNAs (left) or an IRF3 sgRNA (right). See Supplementary Tables 5–7 for more details. (c) Validation of RNA-seq results by RT-qPCR assays of RNA extracted from noninfected vs. HAV-infected PH5CH8 cells. Scatter plots show the ratio of abundance of indicated genes between IRF1 vs. control sgRNA-expressing PH5CH8 cells in HAV-infected (y-axis) and noninfected cells (x-axis). (d) Heatmap showing the relative abundance of indicated genes in noninfected PH5CH8 cells determined by RT-qPCR. (e) Relative HAV RNA abundance on 5 d.p.i. of PH5CH8 transfected with siRNA targeting different IRF1 effector genes. **p<0.01 vs. control. (f) Independent validation of the siRNA results and the combination of 4 siRNAs. *p<0.05, **p<0.01 vs. control. (g) Relative GLuc activity on 3 d.p.i. of HCV-infected Huh-7.5 cells. *p<0.05, **p<0.01 vs. control. (h) Independent validation of the siRNA results and the combination of 3 siRNAs. *p<0.05, **p<0.01 vs. control. (i) Relative DENV RNA on 24 h p.i. of infected Huh-7.5 cells. *p<0.05 vs. control. (j) Independent validation of the siRNA results and the combination of 2 siRNAs. *p<0.05, **p<0.01 vs. control. (k) ZIKV RNA abundance on 24 h p.i. of infected Huh-7.5 cells. *p<0.05, **p<0.01 vs. control. (l) Independent validation of the siRNA results and the combination of 2 siRNAs. **p<0.01 vs. control. Data are means ± s.d. from 3 independent experiments (e-g, i-l) or from 3 technical replicates representative of 2 independent experiments (h). p-values were derived using one-way ANOVA with Dunnett’s multiple comparisons test (e, g, i-l) or two-sided unpaired t-test (f, h). The precise p-values are shown in Supplementary Table 9.