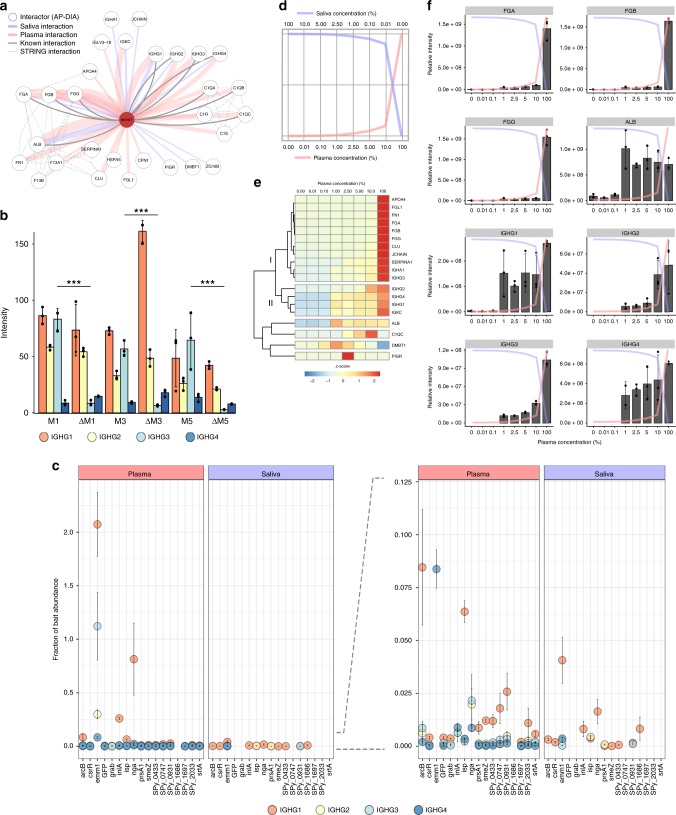
Fig. 5.
The M protein immunoglobulin interactomes. a The M1 protein–human interactome. Known human–human interactions from the STRING-database70 are depicted with broken gray lines. Previously identified M1–human interactions are depicted with solid gray lines, and are most recently described in Hauri et al.17. The interactome views were generated using Cytoscape69 and modified in Adobe Illustrator. b IgG subclass distribution between the different M protein serotypes and their respective deletion mutants. IgG3 is the only subclass for which the difference in binding to the wild-type strain as compared to the mutant strain is significant, as indicated with P values: *P < 0.05, **P < 0.01, and ***P < 0.001 using the Welch’s t-test. c IgG subclass distribution over the different streptococcal bait proteins used in this study. As is evident, the immunoglobulins have a pronounced affinity for the M protein (emm1). The dominating subclass interacting with the M1 protein is IgG1 (orange sphere) followed by IgG3 (shown in light blue). d–f AP–DIA in a mixed plasma–saliva environment mimicking vascular leakage. d Different mixtures of plasma in saliva (0.01%, 0.1%, 1%, 2.5%, 5%, and 10% plasma) were used to study the effect of the host environment on protein–protein interactions. e Cluster analysis of the M1 protein AP–DIA data in a mixed plasma and saliva environment based on z-score. f AP–DIA interaction patterns of fibrinogen (FGA, FGB, FGG), albumin (ALB) and the different IgG subclasses (IGHG1–4) in mixed plasma and saliva environments. Error bars are expressed as standard deviation (s.d.) from the mean. All experiments were prepared using n = 3 biologically independent samples. Source data are provided as a Source Data file