Figure 6.
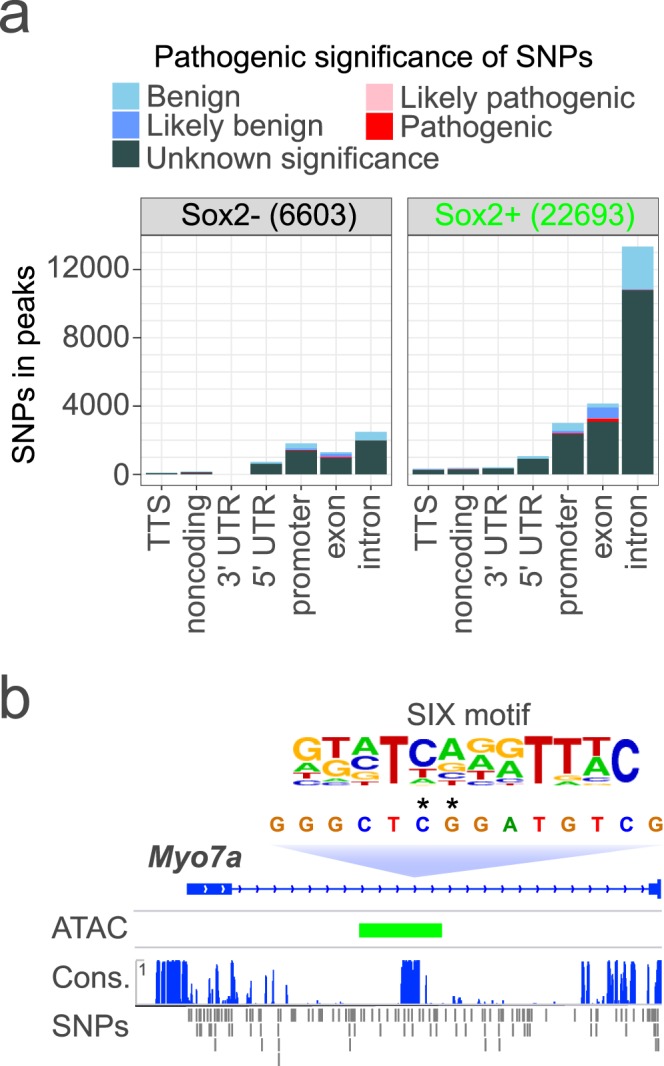
Human deafness gene SNPs in orthologous regions to ATAC-seq peaks detected in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. Numbers of SNPs in the Deafness Variation Database (DVD) overlapping regions orthologous to ATAC-seq peaks in the indicated genomic features are shown in (a). Colors indicate pathogenicity as classified in the DVD. In (b), asterisks indicate two SNPs of unknown significance in MYO7A intron 1 that potentially affect binding at a SIX motif in a region of high evolutionary conservation (Cons.) that is orthologous to an ATAC-seq peak (ATAC) detected in Myo7a in embryonic mouse Sox2-EGFPhigh+ cochlear duct cells.
