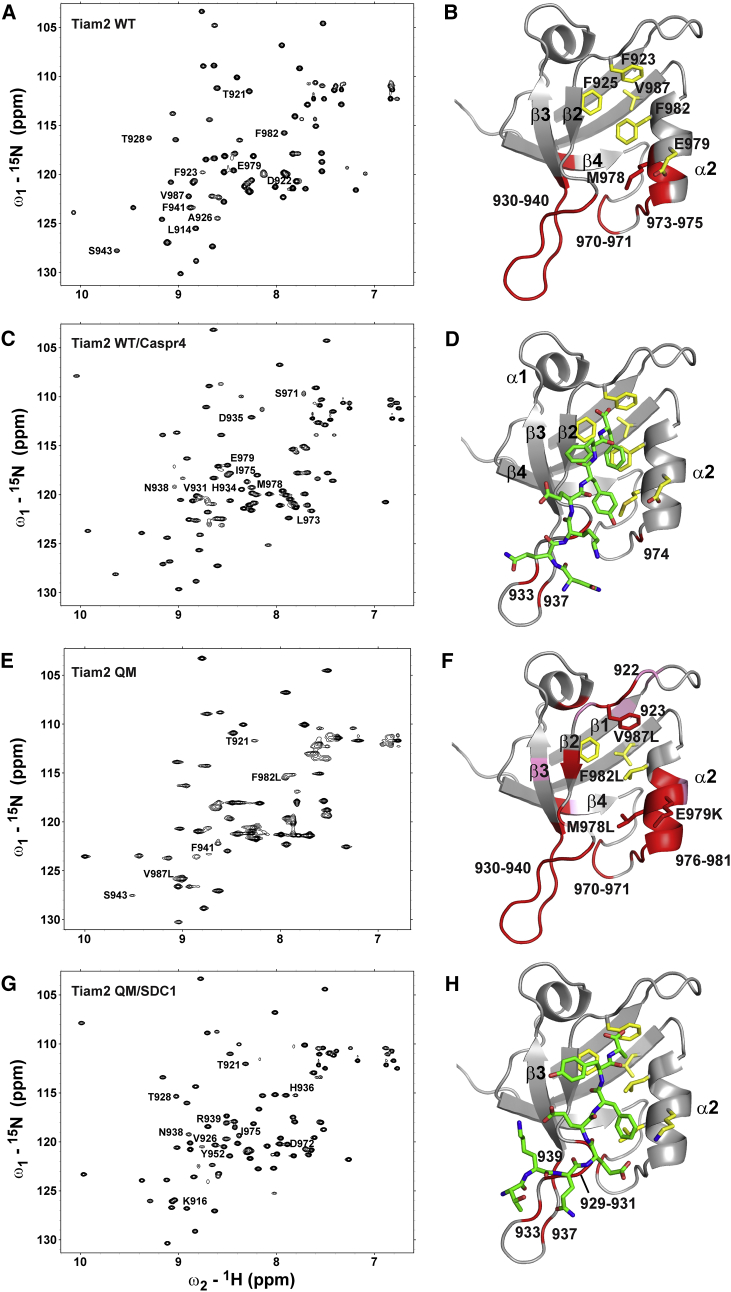
Figure 3.
1H-15N HSQC spectra and structural models of WT and QM Tiam2 PDZ domain free and bound to ligands. (A and B) WT PDZ Tiam2 PDZ in ligand-free state is shown. (C and D) Tiam2 WT PDZ bound to Caspr4 is shown. (E and F) Tiam2 QM PDZ in ligand-free state is shown. (G and H) Tiam2 QM PDZ bound to SDC1 is shown. Each panel represents an HSQC spectrum and the backbone resonance assignments mapped onto the structural model. Select residues are labeled to guide the comparison between spectra. Residues broadened beyond detection are colored in red, whereas residues with weak intensities are colored in pink. The Tiam2 QM homology model was generated by mutating the four residues in the WT homology model created in SWISS-MODEL (26). The PDZ/peptide complexes were generated starting with the apo Tiam2 WT PDZ. The Tiam2 WT/Caspr4 complex was generated by overlaying the Tiam2 WT model with the Tiam1 QM/Caspr4 structure (PDB: 4NXQ) and including the Caspr4 pose with the Tiam2 WT model. The Tiam2 QM/SDC1 complex was generated by overlaying the QM model with Tiam1 WT/SDC1 structure (PDB: 4GVD), keeping the SDC1 pose. These models are shown simply to indicate the location of the peptide-binding site. To see this figure in color, go online.