Figure 1.
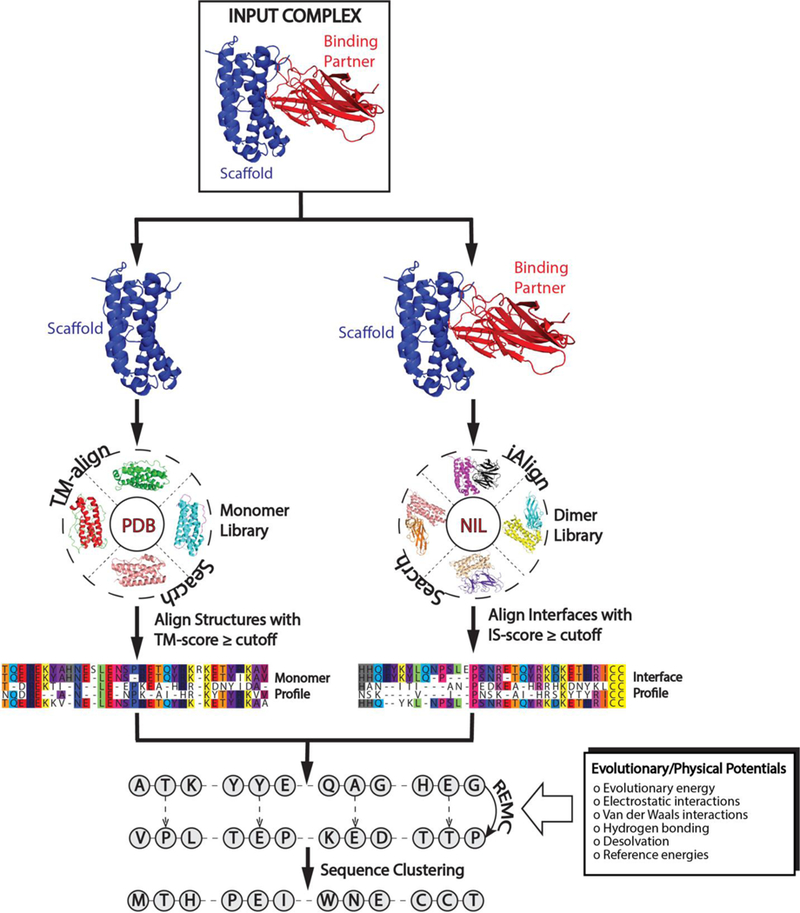
Flowchart of the EvoDesign pipeline for PPI design. Starting from a given protein complex, similar monomer and interface structures are identified from monomer and dimer structure libraries for the scaffold and protein complex, respectively. Alignments of the structural analogies are used to create evolutionary profiles. These profiles are used as energy terms in conjunction with a physical energy function, EvoEF, to guide the replica-exchange Monte Carlo simulation. After clustering the sequence decoys generated during the simulation, the final designs are selected from the lowest free energy sequences in the largest clusters.
