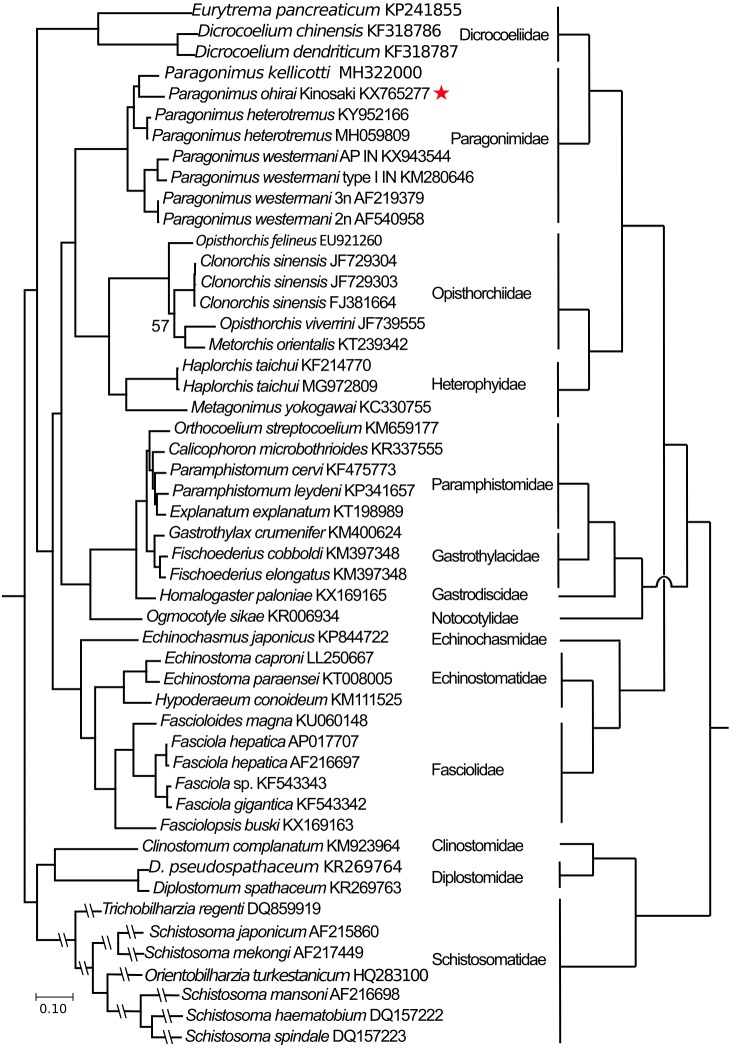
Figure 3. Phylogenetic tree based on concatenated amino-acid sequence data for the 12 mitochondrial proteins from 50 strains of 42 digenean trematode species (Table S1).
Phylogenetic reconstruction was performed in MrBayes 3.2 program (Ronquist et al., 2012) with a final alignment of 2,509 amino-acid residues in length after removing poor-quality regions of the alignment. The tree produced (left-hand side of figure) was a 50% majority-rule tree to which all compatible groupings were added and rooted by outgroup (i.e., members of the Diplostomata –schistosomes and related taxa). Bayesian posterior support values for each node were 100% in every case except one (within the Opisthorchiidae, value of 57% indicated). Accession numbers are given at the end of each sequence label. Paragonimus ohirai in this study is marked by a star symbol. Full names of each species and of families where they belong are provided. The scale bar represents the number of substitutions per site. The tree on the right-hand side of the figure shows the relationships of the same families according to Olson et al. (2003).