Fig. 1. BiP domain architecture and binding cycle.
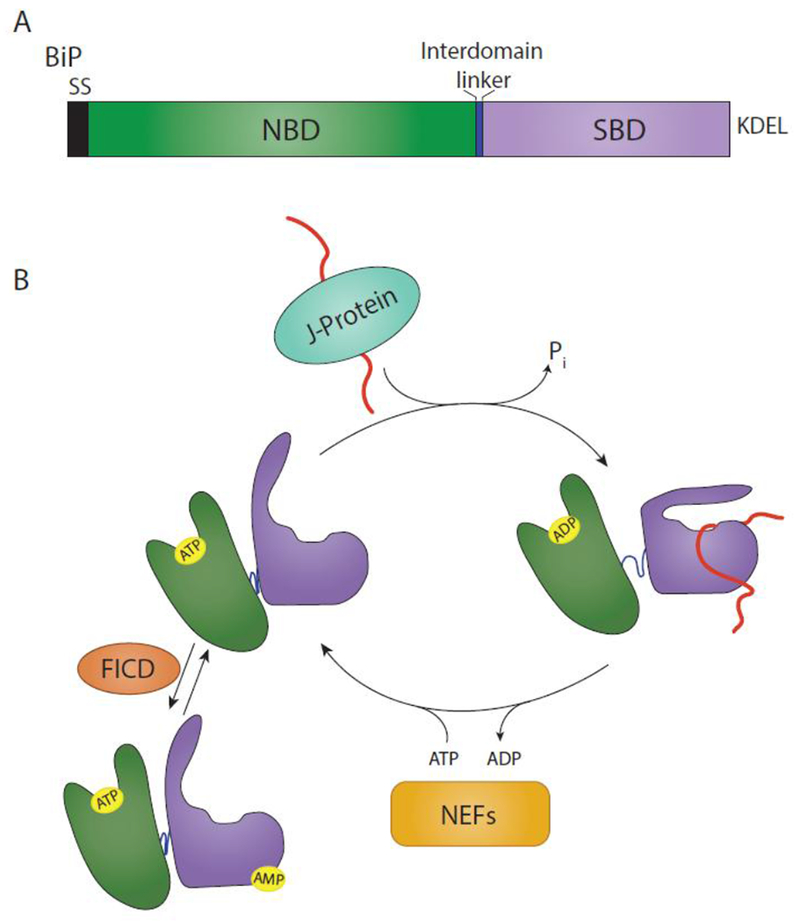
(A) BiP is targeted to the ER via a signal sequence (SS) that is cleaved in the mature form of the protein. From N- to C-terminus, BiP is comprised of a nucleotide binding domain (NBD) (green), interdomain linker (blue), and substrate binding domain (SBD) (purple). It is retained in the ER via a KDEL motif. (B) The substrate binding cycle of BiP is regulated by ATP. When the NBD is bound to ATP, BiP is in a low substrate affinity state. Interaction with a substrate bound J-protein promotes ATP hydrolysis, leading to an extended conformation of the interdomain linker, SBD lid closing and a high substrate affinity. A BiP nucleotide exchange factor (NEF) can then exchange ADP for ATP, placing BiP back in a low substrate affinity state. This process can be inhibited by the AMPylation of BiP by FICD. AMPylation places BiP in a state similar to an ATP-bound state.
