Fig. 3. The calnexin/calreticulin substrate binding cycle.
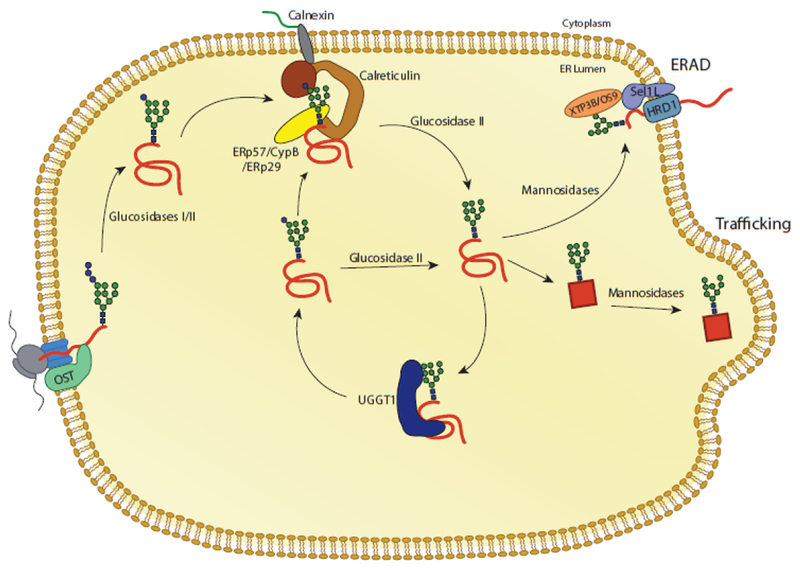
Proteins targeted to the ER receive N-linked glycans that are transferred by the OST complex to acceptor sites. The first two glucoses are trimmed by glucosidases I and II, leaving a monoglucosylated glycan. In this state, the glycan is a substrate for calnexin and calreticulin. Release from calnexin/calreticulin and trimming of the final glucose by glucosidase II leaves the glycan in a non-glucosylated state. Productive folding and adoption of a native state allows for trafficking of the glycoprotein from the ER. Glycoproteins that do not adopt a native fold can be recognized by the folding sensor UDP-glucose: glycoprotein glucosyltransferase 1 (UGGT1). UGGT1 reglucosylates substrates, allowing for rebinding to calnexin/calreticulin or trimming by glucosidase II. Glycoproteins that continue to non-productively fold can be removed from the calnexin/calreticulin cycle through trimming by mannosidases and targeting to ER associated degradation (ERAD) machinery.
