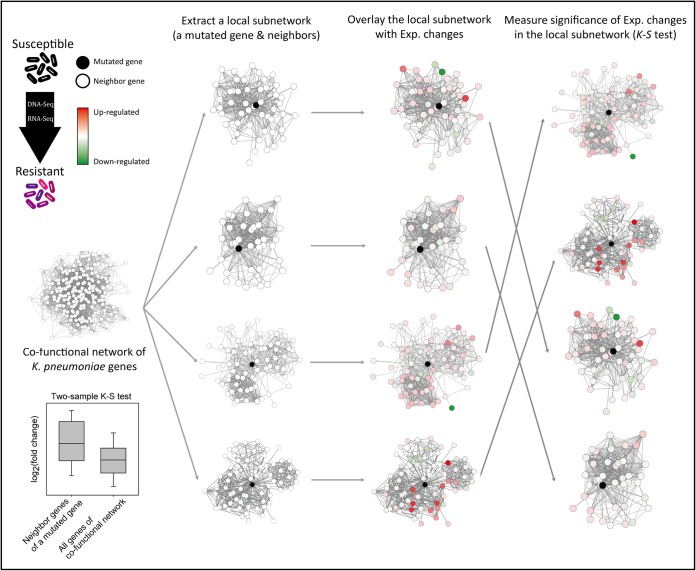
FIG 1.
Schematic overview of network-based prioritization of mutated genes of clinical isolates for antibiotic resistance. Based on the DNA and RNA sequencing profiles of the isolated resistant strains and a susceptible parent strain, we identified genetic alterations and expression (Exp.) changes for all K. pneumoniae genes. Using a cofunctional network of K. pneumoniae genes, we extracted local subnetworks (a mutated gene and its neighbor genes) and overlaid them with expression fold changes. We then measured the significance of the expression changes [log2(fold change)] for network neighbors of each mutated gene by the use of the two-sample Kolmogorov-Smirnov (K-S) test. The four examples of local subnetworks given are ordered from the top row by the P values determined by the K-S test.