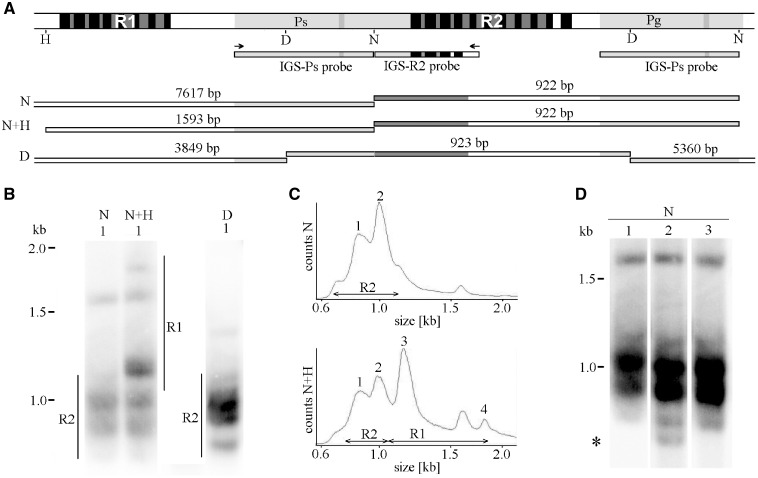
Figure 4.
Size polymorphisms at R2(R1) region(s) may be supported on a genome-wide range by Southern blot analyses. (A) Target(s) for IGS hybridization probe(s) are shown on the background of a restriction map for NdeI (N), HpaI (H) and DraI (D) constructed from the consensus sequence for the corresponding IGS regions (Supplementary Fig. S4). The restricted DNA fragments that are expected to hybridize with the probe(s) are drawn below. (B) Southern hybridization of IGS probe(s) to genomic DNA restricted by the quoted RE reveals multiple rather diffuse bands. For better resolution, only short (<2 kb) fragments are shown (long fragments are shown in Supplementary Fig. S10). (C) Densitometric evaluations of hybridization profiles. Note that NdeI digests show size variability in R2 (Bands 1–2), whereas double digestion with NdeI and HpaI shows size variability in both R1 (Bands 3–4) and R2 (1–2) regions. (D) Longer exposition of NdeI digests highlighted structural variability (asterisk) across three plants selected within a single pumpkin cultivar. Plant marked by number 1 was sequenced and analysed in panel (B).