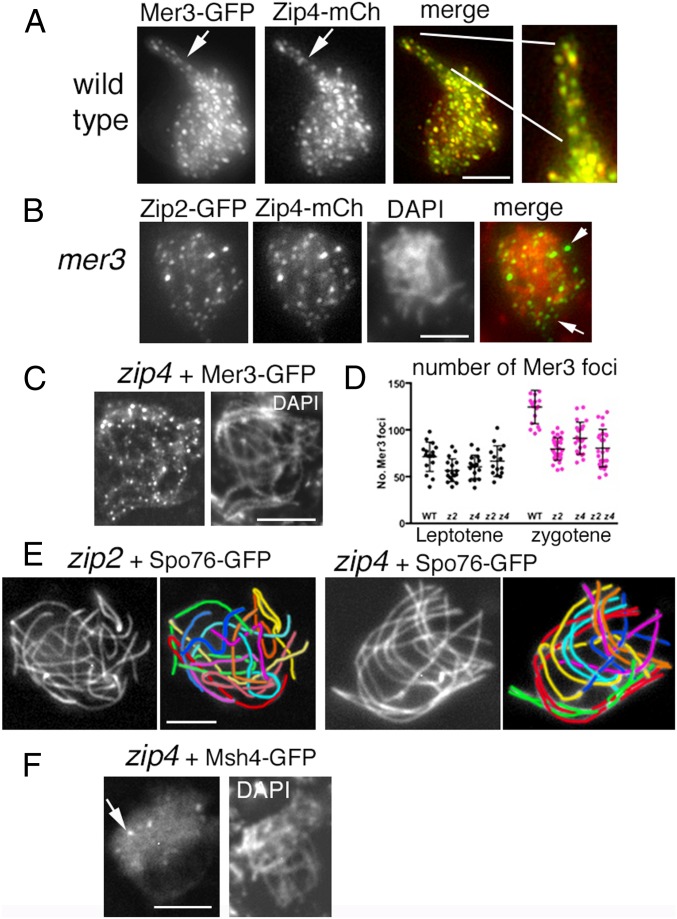
Fig. 5.
Zip2-Zip4, Mer3, and Msh4 localizations and zip2-zip4 pairing defects. (A) Zip4 localizes to the matching pairs of Mer3 foci in WT. (A, Right) Enlarged segment of the coaligned region. (B) Zip2 and Zip4 colocalize in the absence of Mer3, but foci are no longer exclusively located on chromosomes, as shown by their merge with DAPI (Right; arrows point to foci which do not overlap with DAPI staining). (C) Mer3 foci are regularly spaced along all zip4 chromosomes. (D) Number of Mer3 foci in single and double zip2 zip4 mutants (z2, z4, z2z4) compared with WT at leptotene (Left, black dots) and at early zygotene (Right, pink dots) nuclei (determined by ascus sizes, which grow from 20 μm at early leptotene to 60 μm at zygotene and 100 μm at pachytene, in mutants and WT). (E) zip2 and zip4 mutant nuclei show either no coalignment at all (Left) or partial coalignment (Right). The seven homologs are distinguishable by their lengths and color in the corresponding drawings. In the zip4 nucleus (Right), red, green, cyan, and orange pairs are coaligned while purple pair is partially coaligned and yellow plus blue pairs are not coaligned. Chromosome axes are marked by Spo76-GFP. (F) Only few Msh4 foci (arrow) are visible in the absence of Zip4 (compare with Mer3 foci in C). (Scale bars: 2 μm.)