Figure 3.
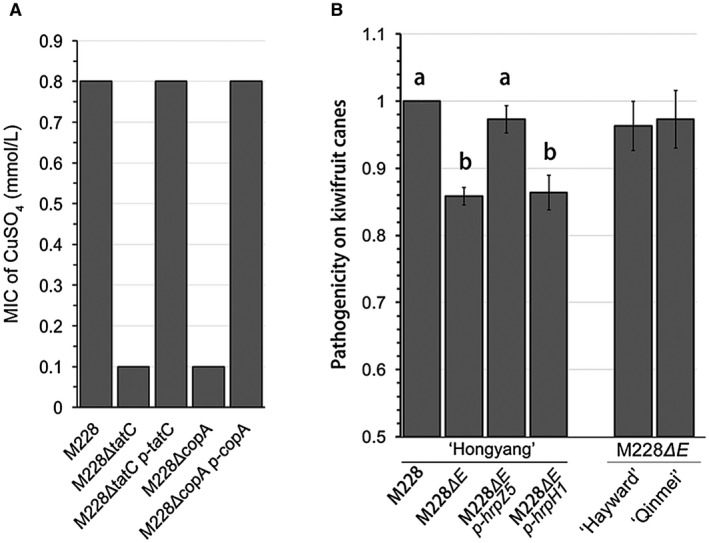
The polymorphic genes copA and hopZ5 revealed by comparative genomics amongst three clades within Pseudomonas syringae pv. actinidiae biovar 3 (Psa3) are required for copper resistance and pathogenicity, respectively. (A) The minimal inhibitory concentration (MIC) of CuSO4 for the tatC and copA mutants of Psa3 determined by the agar‐dilution method. The LB plates contained eight twofold serial dilutions of CuSO4 with a starting concentration of 3.2 mM. An in‐frame deletion of either the copA or tatC gene resulted in increased susceptibility to CuSO4, while the changed phenotype could be fully complemented by expressing the corresponding genes. (B) HopZ5 was required for the full virulence of Psa3 during infection. An in‐frame deletion of the hopZ5‐hopH1 cluster from M228 resulted in M228ΔE, which showed significantly reduced pathogenicity on canes of A. chinensis var. chinensis ‘Hongyang’, but not on canes of A. chinensis var. deliciosa ‘Hayward’ and ‘Qinmei’. The reduced pathogenicity could be restored by expressing hopZ5 (M228ΔE p‐hrpZ5), but not hopH1 (M228ΔE p‐hrpH1). The lesion length measured at 15 days post‐inoculation (dpi) of each mutant was normalized to that of M228. The experiments were repeated three times with similar results.
