FIGURE 4:
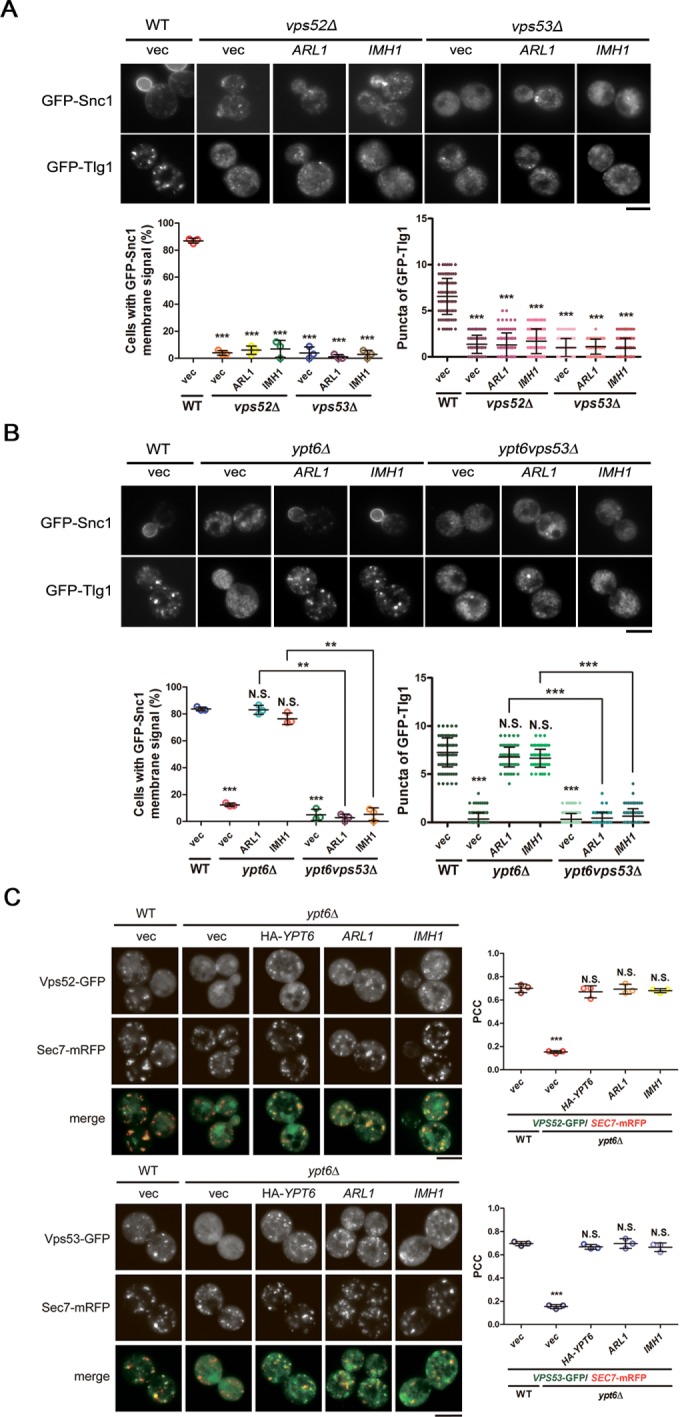
The recruitment of the GARP complex to the TGN by Arl1 and Imh1 is critical for suppressing defects in endosome-to-TGN transport in ypt6Δ cells. (A) Overexpression of Imh1 or Arl1 is unable to suppress GFP-Snc1 and GFP-Tlg1 mislocalization in GARP-complex subunit-deletion strains. Arl1 or Imh1 was coexpressed with GFP-Snc1 or GFP-Tlg1 in vps52Δ and vps53Δ cells. Live cells were observed in mid–log phase by fluorescence microscopy. Cells showing the GFP-Snc1 plasma membrane signal or the GFP-Tlg1 puncta were quantified (n = 100). The data are reported as the mean ± SD of three independent experiments (***p < 0.001; one-way ANOVA; scale bar, 5 μm). (B) The GARP complex is required for Arl1 and Imh1 to suppress Snc1 and Tlg1 transport defects in ypt6Δ cells. Fluorescently tagged Snc1 or Tlg1 was coexpressed with Arl1 or Imh1 in ypt6Δ or ypt6vps53Δ cells. Live cells were observed in mid–log phase using fluorescence microscopy. Cells showing the GFP-Snc1 plasma membrane signal or the GFP-Tlg1 puncta were quantified (n = 100). The data are reported as the mean ± SD of three independent experiments (**p < 0.005, ***p < 0.001; N.S. not significant; one-way ANOVA; scale bar, 5 μm). (C) Overexpression of Arl1 or Imh1 restores the late Golgi puncta of the GARP complex in ypt6Δ cells. The Arl1-, Imh1-, or HA-Ypt6-overexpressed ypt6Δ cells were coexpressed with fluorescently tagged Vps52 or Vps53 and Sec7-mRFP (late Golgi marker). Live cells were observed in mid–log phase using fluorescence microscopy (scale bar, 5 μm). At least 100 cells were included in the experiments from three independent biological repeats. The ratios of colocalization between Vps52/53-GFP and the late Golgi indicator (Sec7-mRFP) were represented as the Pearson correlation coefficient (PCC) using the ImageJ plug-in Just Another Colocalization Plugin with Costes Automatic Thresholding as described in Materials and Methods. The data are presented as the mean ± SD, and the p values were determined applying one-way ANOVA (***p < 0.001; N.S. not significant).
