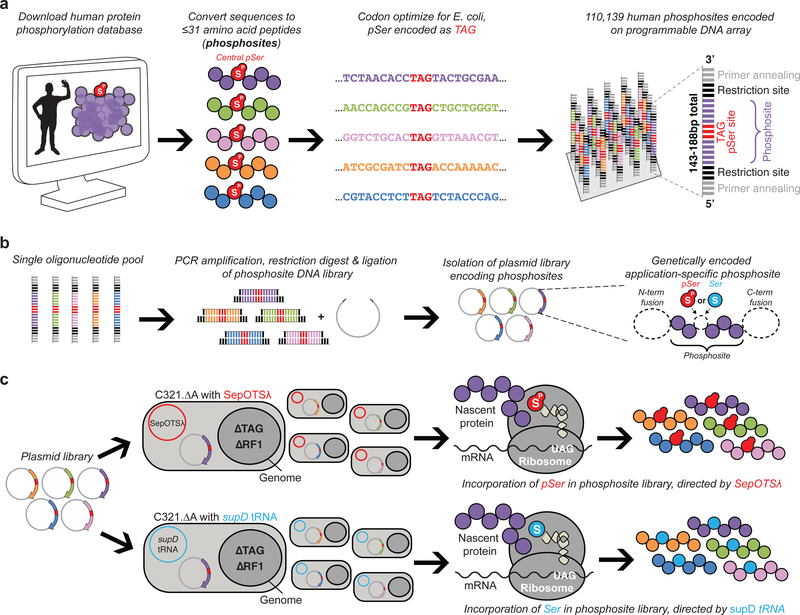
Figure 1: Design and display of the synthetic human serine phosphoproteome.
(a) Recombinant human phosphosite DNA sequences were designed based on previously-observed instances of serine phosphorylation from the PhosphoSitePlus database11 and synthesized as oligonucleotides harboring a central TAG codon to direct pSer or Ser incorporation. The 16–31 amino acid phosphosites including the TAG codon were encoded as 48–93 bp oligonucleotides, and additional restriction and primer annealing sites were added to both ends, yielding 143–188 bp sequences. (b) All oligonucleotide sequences encoding phosphosites were liberated from the microarray, PCR-amplified in a single pool, restriction digested, and introduced into an application-dependent expression vector. (c) The phosphosite-encoding plasmid library was then transformed into genomically recoded E. coli (C321.ΔA) lacking all endogenous UAG codons and release factor 1 (RF1), which normally terminates translation at UAG codons. The library was separately transformed into C321.ΔA strains containing either a translation system to insert pSer (SepOTSλ) or Ser (supD tRNA) at UAG codons, enabling the synthesis of either the phosphorylated or unphosphorylated version of the phosphosite library. This workflow was employed for various applications of the phosphosite library, as dictated by the expression vector used for experimentation.