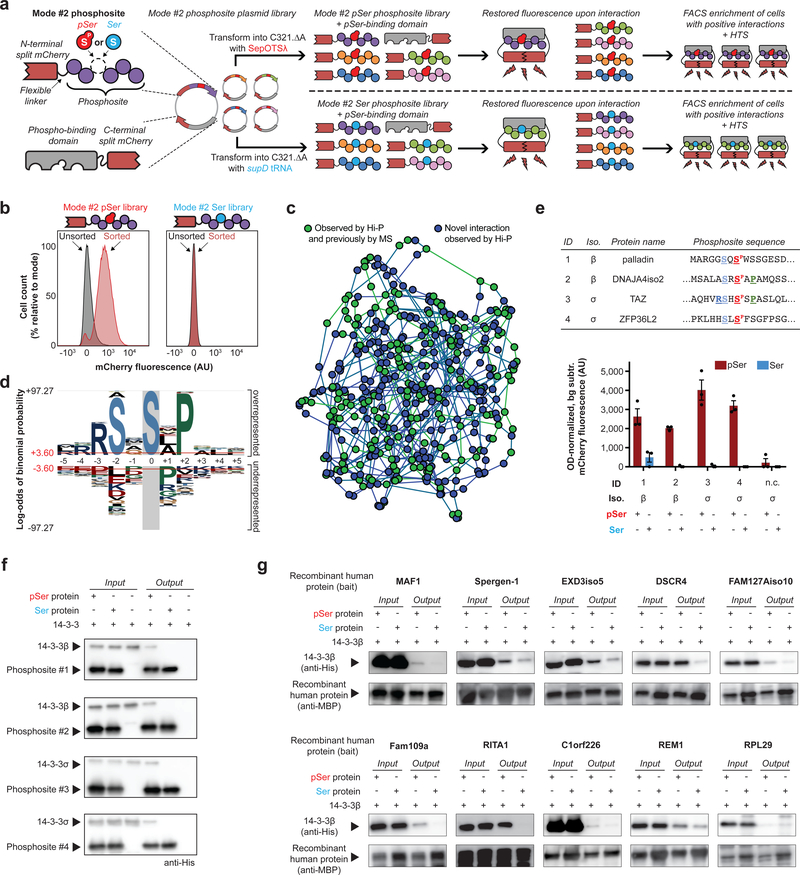
Figure 3: Identification of pSer-dependent protein interactions with 14–3-3 isoforms by Hi-P.
(a) Hi-P experimental workflow. Split mCherry in E. coli enables identification of protein interactions by restored fluorescence signal. Cells expressing mode #2 phosphosites that interact with a phospho-binding protein are isolated by FACS, and implicated phosphosites are identified by HTS. This scheme can be performed with either SepOTSλ or supD tRNA to study pSer- or Ser-containing mode #2 phosphosites, respectively. (b) Hi-P experiments with 14–3-3β yielded increased mean population fluorescence after FACS with SepOTSλ (encoding pSer) but not with supD tRNA (encoding Ser). n=105 cells for flow cytometry observation. (c) A network illustrating the relationships amongst the Hi-P hits using 14–3-3β and SepOTSλ. Each node represents a phosphosite sequence identified by Hi-P and is connected to the two phosphosite nodes closest in sequence space by weighted edges. Green nodes indicate the phosphosite is derived from a protein that has been identified as a 14–3-3 interactor in previous mass spectrometry (MS) experiments17 and blue nodes are novel candidate interactions identified via Hi-P. (d) pLogo analysis19 of 14–3-3β Hi-P results (n = 388 phosphosites above 1,000 HTS read cutoff identified from a single Hi-P experiment). Significance calculated by binomial probability of amino acid frequencies, red lines indicate p = 0.05 significance threshold with Bonferroni correction. (e) Validation of select 14–3-3 Hi-P hits by BiFC shown in bar graph. Top-ranking phosphosite sequences were identified by Hi-P using either 14–3-3β or 14–3-3σ isoforms, as indicated. Amino acids surrounding the central pSer residue (in red) adhering to the RSXSPXP motif are colored and bolded. Background fluorescence for isogenic cells in which mode #2 phosphosite expression was not induced was subtracted, and fluorescence was normalized by OD600. Error bars show s.e.m. centered at mean (n = 3 independent replicates); n.c. = negative control phosphosite not anticipated to interact with 14–3-3 proteins AGPADAPAGAVVGGG[SP/S]PRGRPGPVPAPGLLA. (f) Pull-down analysis of immobilized mode #1 phosphosites confirmed pSer incorporation is necessary for 14–3-3 interaction, representative of two independent replicates. (g) Pull-down analysis of immobilized MBP-fusion full-length recombinant human phosphoproteins confirmed pSer is necessary for or enhances 14–3-3β interaction for 9/10 of the tested proteins, representative of two independent replicates. AU, arbitrary units.