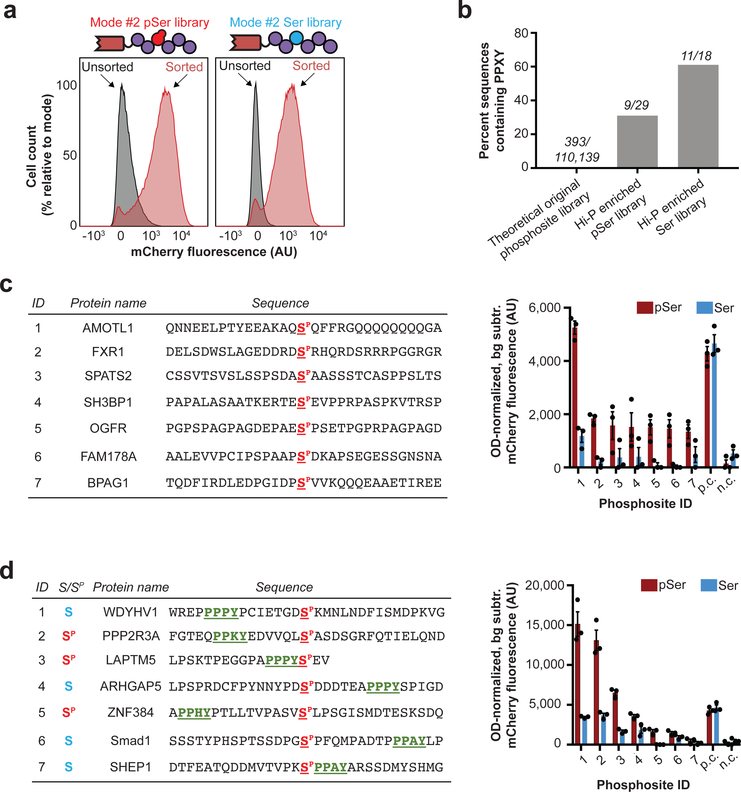
Figure 4: Investigation of NEDD4 WW2 interactions by Hi-P.
(a) Hi-P experiments with cells co-expressing the NEDD4 WW2 domain and the mode #2 phosphosite library yielded increased mean population fluorescence with either SepOTSλ (encoding pSer) or supD tRNA (encoding Ser). n=105 cells for flow cytometry observation. (b) Hi-P experiments using WW2 from NEDD4 resulted in enrichment of PPXY-containing phosphosites in both pSer- and Ser-encoding populations. The raw number of sequences containing PPXY over number of sequences in population are shown above each bar. All data is for a 1,000-read cutoff by HTS. (c) BiFC analysis of select NEDD4 WW2 Hi-P phosphosite hits from SepOTSλ (encoding pSer) experiments and excluding sequences containing the PPXY motif. (d) BiFC analysis of select NEDD4 WW2 Hi-P phosphosite hits, with PPXY motif (green underlined), from experiments with either SepOTSλ (encoding pSer) or supD tRNA (encoding Ser). For BiFC experiments in (c) and (d), background fluorescence for isogenic cells in which mode #2 phosphosite expression was not induced was subtracted, and fluorescence was normalized by OD600. Error bars show s.e.m. centered at mean (n = 3 independent replicates). n.c., negative control mode #2 peptide WFYSPFLE co-expressed with mouse Nedd4 WW2 fused to C-terminal split mCherry22; p.c., positive control mode #2 peptide IPGTPPPNYD co-expressed with mouse Nedd4 WW2 fused to C-terminal split mCherry22; AU, arbitrary units.