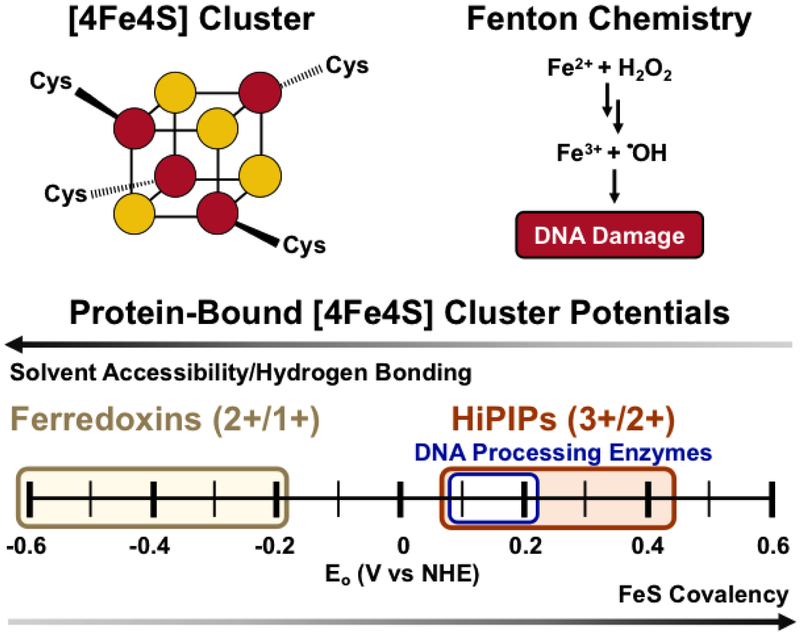
Figure 1. [4Fe4S] Clusters.
(Above, left) Schematic of the cubane [4Fe4S] cluster, with Fe in red and S in yellow. (Above, right) Cofactors containing iron can react with cellular oxidants leading to Fenton chemistry and DNA damage by the hydroxyl radical. (Below) The potential of the [4Fe4S] cluster cofactor is tunable over a wide range of physiological redox potential values. Ferredoxins access the [4Fe4S]2+/1+ couple, upon reduction from the resting [4Fe4S]2+ state. (yellow). High potential iron proteins (HiPIPs) access the [4Fe4S]3+/2+ couple upon oxidation to the [4Fe4S]3+ state from the resting [4Fe4S]2+ state (red). DNA-processing enzymes with [4Fe4S] cofactors have DNA-bound redox potentials which fall within the HiPIP [4Fe4S]3+/2+ potential range, at approximately ~65-150mV vs. NHE (blue). The solvent accessibility and hydrogen bonding/electrostatic environment of the cluster all contribute to tuning the redox potential of the cofactor (4,5).