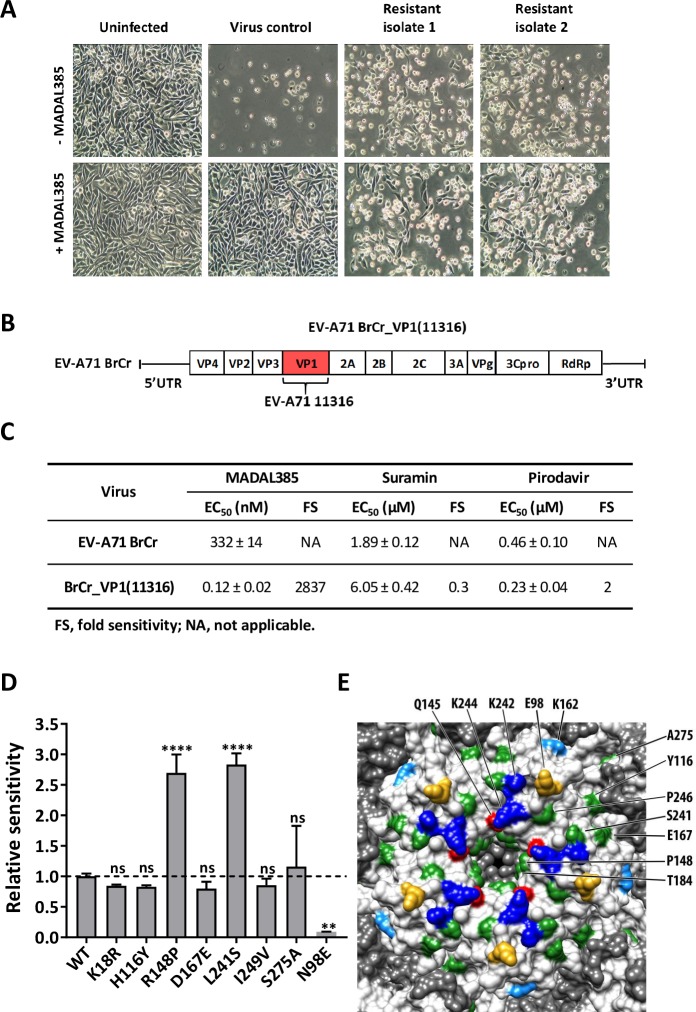
Fig 2. Selection and characterization of MADAL385-resistant and -sensitive EV-A71 strains.
(A) EV-A71 BrCr and resistant strains were used to infect RD cells in presence or absence of MADAL385 (3 μM). Images were taken at 1-day post infection. (B) Schematic representative of the recombinant EV-A71 BrCr_VP1(11316). (C) In vitro EC50s of MADAL385, suramin and pirodavir against EV-A71 BrCr and EV-A71 BrCr_VP1(11316) infection in RD cells. (D) Relative sensitivity of 8 EV-A71 single mutants to MADAL385. Data were normalized on the EC50 of EV-A71 BrCr (WT). Statistical analysis was performed using the one-way ANOVA test relative to the wild-type; **p<0.01; ****p<0.0001; ns, not significant. (E) Surface rendering of the EV-A71 pentamer was generated using USCF Chimera (PDB ID: 3VBS). VP1 residues that bind PSGL1 and HS (K242 and K244, blue) or determine the receptor-binding phenotype (Q145, red) or identified as HS-binding (K162, light blue), or complementing HS binding (E98, gold) are colored accordingly and indicated (top). Positions associated with MADAL385 resistant/sensitive phenotype are colored in green and indicated (right). Amino acids were labeled according to the corresponding residues of EV-A71_11316. Error bars represent the mean ± SD of at least 2 independent experiments with 3 independent replicates each.