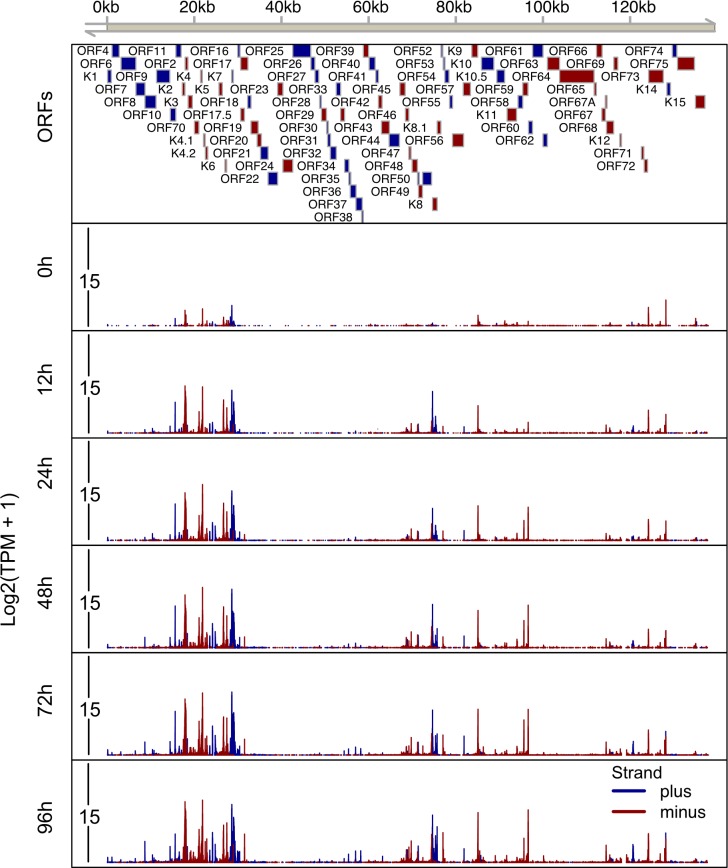
Fig 3. High resolution mapping of transcription start sites on the KSHV genome in TREx-BCBL1-RTA cells.
Total RNA was isolated from TREx-BCBL1-RTA cells at 0h, 12h, 24h, 48h, 72h and 96h post-dox induced lytic reactivation and RAMPAGE libraries were prepared and sequenced. The 5’ signal of read 1, which corresponds to transcriptional start sites, were mapped on the KSHV genome (GQ994935.1). A schematic of the KSHV genome including genomic coordinates of annotated ORFs is depicted on the top horizontal axis. The lower 6 tracks transcripts initiated from every nucleotide across the whole KSHV genome at each time point. The Y axis depicts the log2 transformed TPM value. Blue boxes/lines indicate ORFs/TSSs on the plus strand, red boxes/lines indicate ORFs/TSSs on the minus strand.