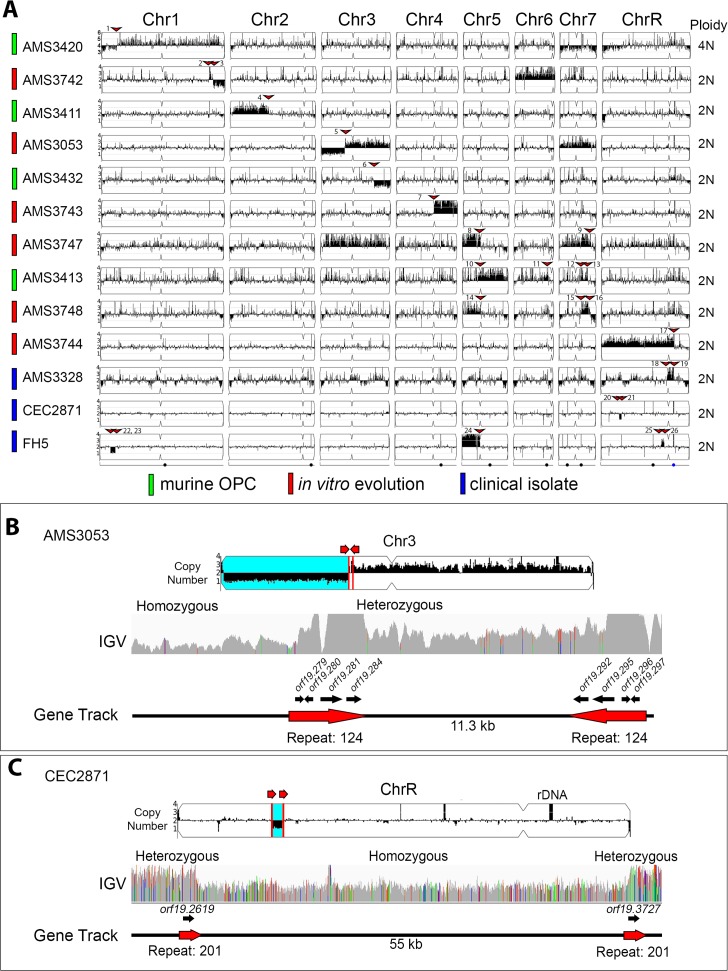
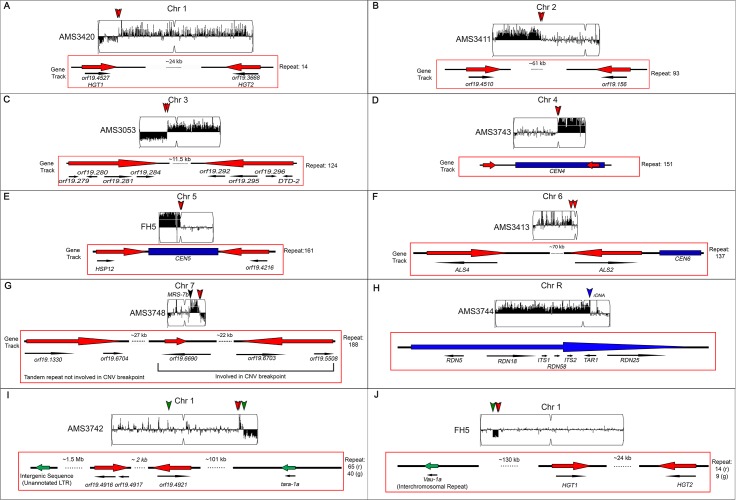
Figure 3. All copy number breakpoints resulting in segmental aneuploidy occur at repeat sequences.
(A) Whole genome sequence data plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP. The source of each isolate is indicated in color: in vivo evolution experiments in a murine model of oropharyngeal candidiasis (OPC) (green), in vitro evolution experiments in the presence of azole antifungal drugs (red), and clinical isolates (blue). Ploidy, determined by flow cytometry, is indicated on the far right. Every copy number breakpoint occurred at a repeat sequence (red arrow), additional details are in Supplementary file 3. Location of the Major Repeat Sequences (black circle) and rDNA array (blue circle) are shown below. Example copy number breakpoints for two isolates (B–C). (B) Isolate AMS3053 underwent a complex rearrangement on Chr3L at a long inverted repeat (Repeat 124, red lines). Read depth (top panel) and allele frequency (IGV panel) data indicate the copy number breakpoint coincided with LOH (blue region) telomere proximal to the breakpoint. The inverted repeat copies (~3.2 kb, 99.5% sequence identity, separated by ~11.3 kb) each contain four complete ORFs and intergenic sequences. (C) Read depth (top panel) and allele frequency (IGV panel) data for isolate CEC2871 shows an internal chromosome deletion on ChrR with copy number breakpoints (red lines) and LOH (blue) that occur between a long tandem repeat (Repeat family 201, red arrows). The tandem repeat copies (~1.4 kb, 93.8% sequence identity, separated by ~55 kb) each contain one ORF.