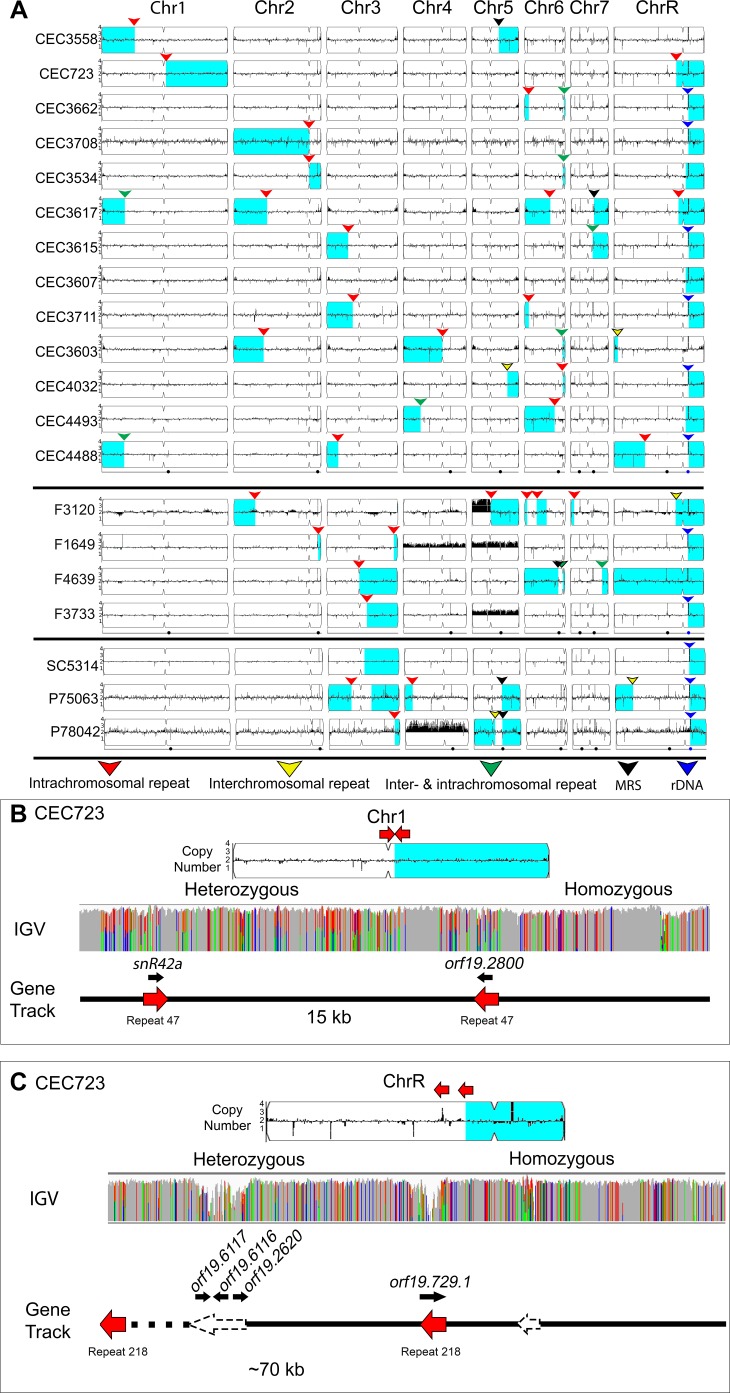
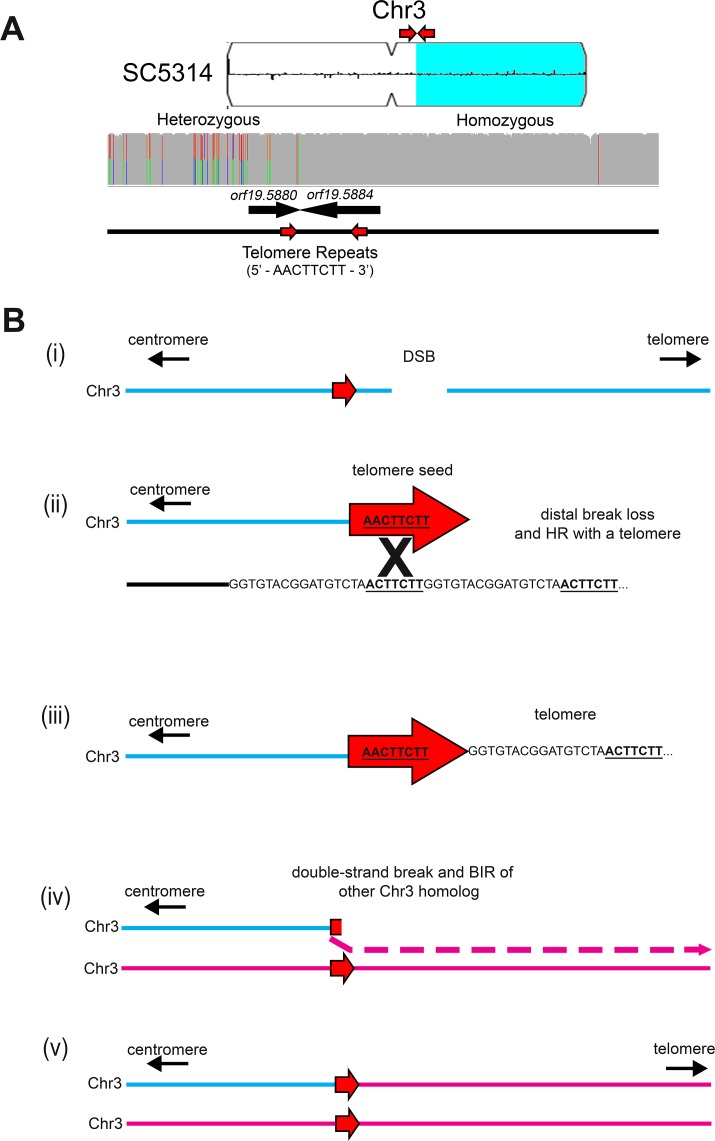
Figure 4. Many LOH breakpoints occur at long intra- and inter-chromosomal repeat sequences.
Whole genome sequence data plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP. (A) All long-range homozygous regions (light blue) that are associated with long repeat sequences (colored arrows) are indicated for 20 diverse C. albicans isolates. LOH breakpoints and isolate information are detailed in Supplementary files 1 and 4. The type of long repeat is indicated with colored arrows: intra-chromosomal (red), inter-chromosomal (yellow), both intra- and inter-chromosomal (green), rDNA repeat (blue), and MRS (black). (B–C) Two example LOH breakpoints in isolate CEC723 that occur at long repeats (red arrows) on (B) Chr1 (repeat copy length ~1.1 kb), and (C) ChrR (repeat copy length ~3.3 kb) and continue to the right telomere of the respective chromosomes. Heterozygous and homozygous allele ratios are indicated in the IGV track. The position, orientation, and spacer length of the long repeat sequence is indicated in the gene track. ORFs (black arrows) contained within the long repeat sequences are indicated above the gene track. The LOH breakpoint on ChrR is within a repeat-dense region; additional long repeats in the region are indicated (dashed arrows).