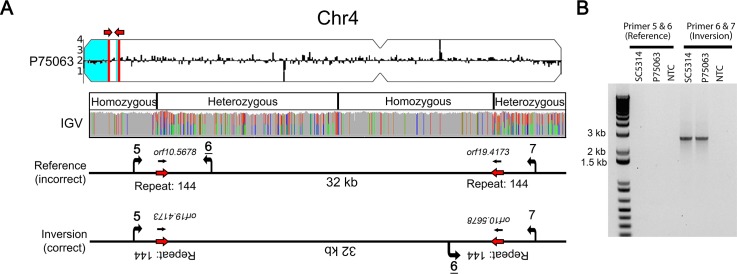
Figure 5. Long repeat sequences are associated with chromosomal inversions.
(A) Whole genome sequence read depth plotted as a log2 ratio and converted to chromosome copy number (Y-axis) and chromosome location (X-axis) using YMAP. Long-range homozygous regions (blue) on Chr4 are indicated for the isolate P75063. IGV allele ratio track indicates multiple homozygous to heterozygous transitions between a long inverted repeat (red arrows, repeat 144, copy length ~1.7 kb). Primers (5, 6, and 7, Supplementary file 7) were designed to unique sequences flanking repeat 144. (B) PCR amplification between Primers 6 and 7 identifies a ~32 kb chromosomal inversion in both the reference isolate SC5314 and P75063; the reference orientation did not amplify (Primers 5 and 6).