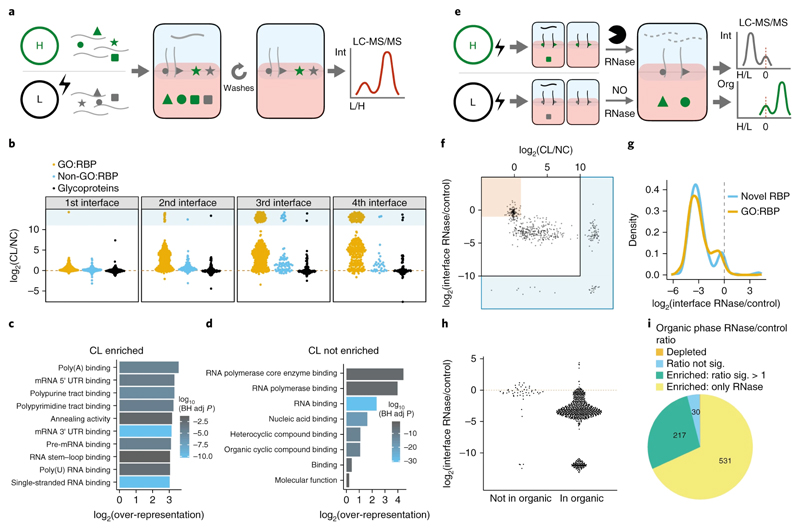
Figure 2. OOPS for RBP recovery.
(a) Schematic representation of the SILAC experiment used to determine the effect of UV crosslinking on protein abundance in the interface and the effect of additional phase separation cycles to wash the interface. Equal quantities of cells +/- UV crosslinking are labelled with SILAC and mixed prior to OOPS. RNA bound proteins are expected to have a positive CL vs NC ratio. Contaminants are expected to be equally abundant in CL and NC.
(b) Protein CL vs NC ratios for the 1st to 4th interfaces. Infinite ratios (not detected in NC) are presented as pseudo-values in blue box. GO:RBP = GO annotated RNA binding protein.
(c) Top 10 molecular function GO terms over-represented in proteins enriched by CL in the 3rd interface. BH adj p-value = Benjamini-Hochberg adjusted p-value. P-value obtained from a modified hypergeometric test to account for protein abundance (see online methods).
(d) As per (c) for proteins not enriched by CL in the 3rd interface.
(e) Schematic representation of the SILAC experiment to determine protein abundance in the 3rd interface and 4th organic phase following RNase treatment. Equal quantities of cells were UV crosslinked and RNA-protein adducts enriched by OOPS +/- RNase before combining the samples for a final phase separation in which both the interface and the organic phase are collected. Proteins from RNase treated cells will be depleted from the interface and enriched in the organic phase.
(f) Protein CL vs NC ratio and RNase vs control ratio in the interface. Red box denotes proteins which are not CL-enriched and not depleted by RNase. The blue regions surrounding the graph denote ratios which cannot be accurately estimated as the protein was only detected in one condition and therefore a pseudo-value is presented.
(g) RNase vs control ratio in the interface for GO annotated RBPs and other OOPS RBPs
(h) Protein RNase vs control ratio in the interfaces for proteins identified in the 4th step organic phase. Red line = equal intensity in RNase-treated and control.
(i) Proportion of proteins enriched in the organic phase following RNase treatment. Proteins detected in both +/- RNase conditions but with insufficient peptides to test for significant enrichment are excluded.