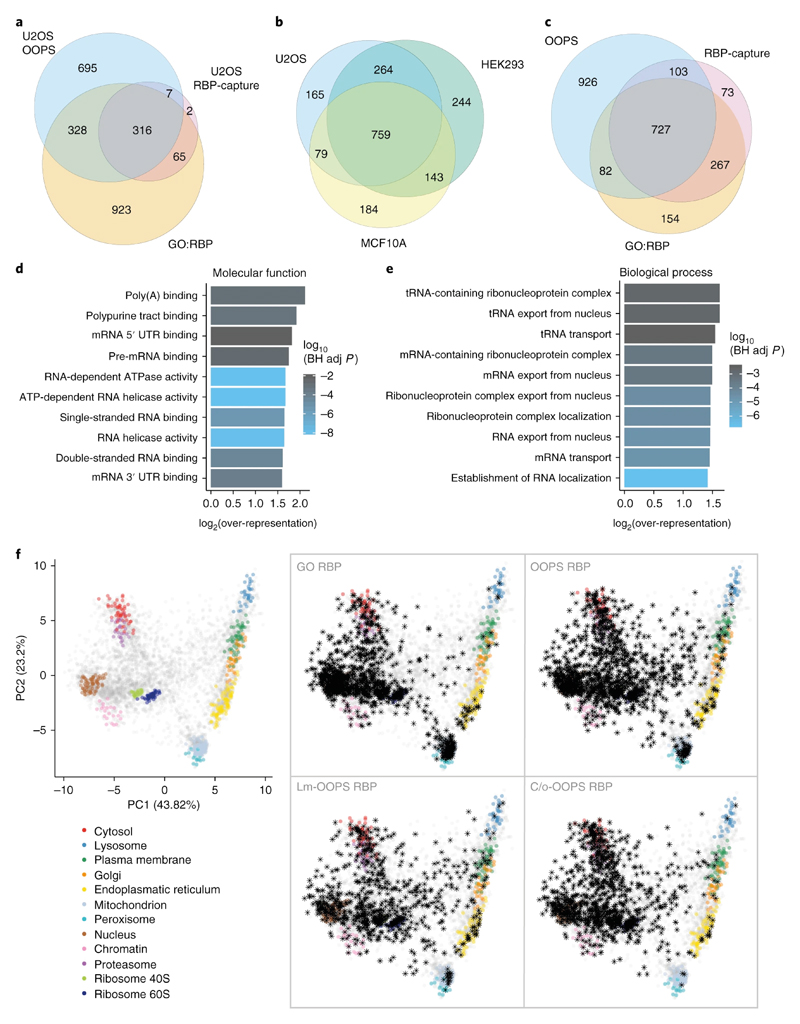
Figure 3. RBPs identified using OOPS.
(a) Overlap between OOPS, RBP-Capture and GO-annotated proteins for U2OS cells. Proteins were restricted to those expressed in U2OS.
(b) Overlap between proteins identified with OOPS from U2OS, HEK293 and MCF10A. Proteins were restricted to those expressed in all cell lines.
(c) Overlap between the union of OOPS RBPs identified in the 3 cell lines in (b), all published RBP-Capture studies, and GO annotated RBPs. Proteins were restricted to those expressed in all 3 OOPS cell lines.
(d) Top 10 molecular function GO terms over-represented in the proteins identified in U2OS OOPS. BH adj p-value = Benjamini-Hochberg adjusted p-value. P-value obtained from a modified hypergeometric test to account for protein abundance (see online methods).
(e) As per (d) for novel U2OS RBPs identified by OOPS.
(f) HyperLOPIT projections of protein steady state localisation. Left: Canonical subcellular localisation markers indicated in colour as shown. Right: Highlighted RBPs shown as black asterisks. GO RBP = GO annotated RBP. Lm = Light membrane-enriched fraction. C/o = Cytoplasm/Other fraction. Annotated proteins in each fraction were detected in at least one of 5 repeat experiments.