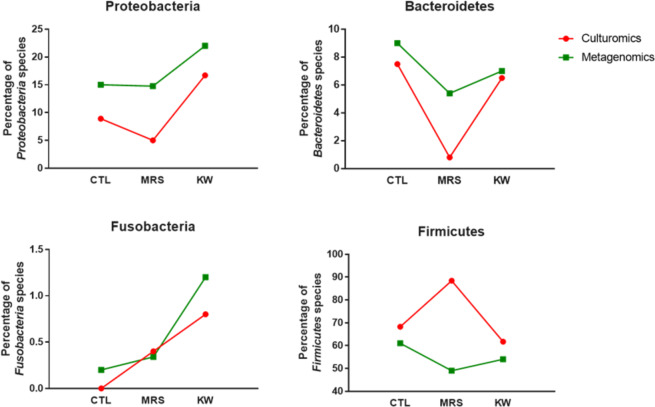
Figure 4.
Phylum distribution in culturomics and metagenomics. Culturomics and metagenomics results confirmed a significant increase in Proteobacteria in kwashiorkor (uncorrected chi-square test, CTL-KW, MRS-KW, p < 0.001) as well as a significant decrease in Bacteroidetes in marasmus (uncorrected chi-square test, CTL-MRS, MRS-KW, p < 0.0005). Strikingly, the increased Fusobacteria diversity in kwashiorkor observed by metagenomics was also confirmed in kwashiorkor, but not in marasmus, by culturomics (uncorrected chi-square test, CTL-KW, p = 0.004).