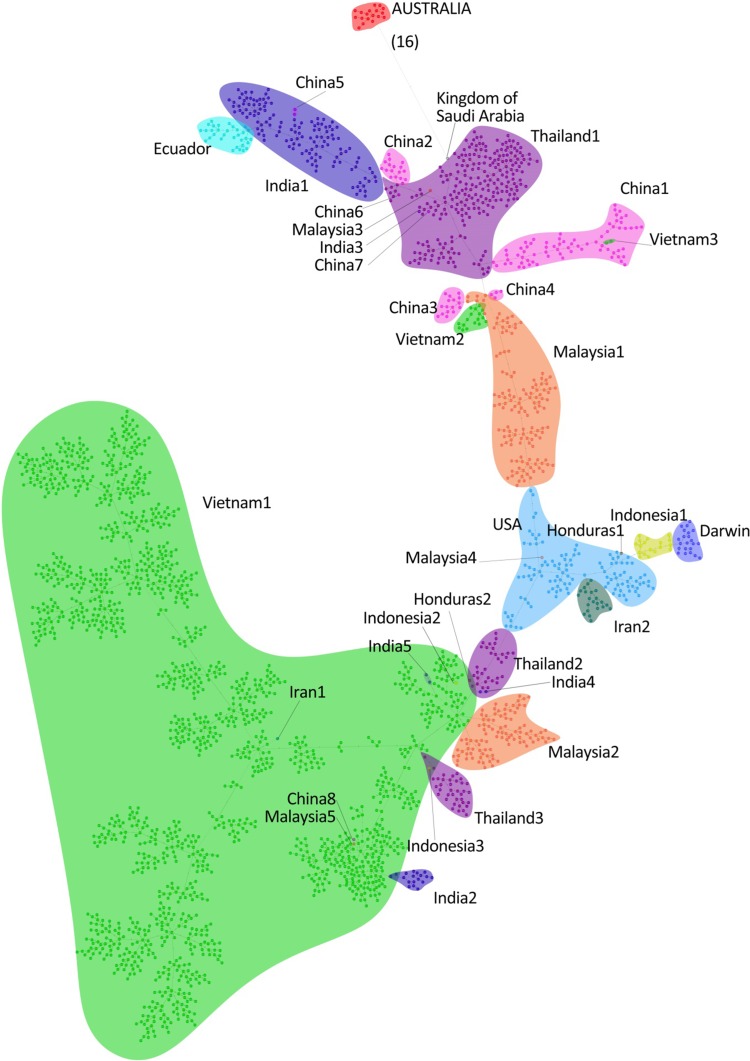
Fig. 1.
Minimum spanning tree of WSSV genotypes (stylised for ease of labelling and navigation). Where there are multiple clusters from the same region, the numerical codes relate to the following samples: China1: C1, C2, C4, C5, C6, C7, C10, C16, C19, C71, C72, C73, C74, C75, IT14. China2: IT5. China3: IT2. China4: IT9. China5: IT6, IT12. China6: IT38. China7: IT44. China8: C30. India1: NAP1, NAP2, NAP3, NTN3, NTN4, NTN5, NWB1, NKE1, NKE2, NKE3, NKE4, NKE5. India2: OTN1, OAP1, OGU1, OWB1. India3: NTN2. India4: OOD1, OKE1. India5: NTN1. Thailand1: T16, T19, T20, T101, T103, T105, T106, T108, T110, T116, T119, T120, T121, T122, T123, T124, T125. Thailand2: ThC-98. Thailand3: ThaiMB, F17, T140, T142, T143. Malaysia1: IT7, IT8, IT10. Malaysia2: IT1, IT11, IT19. Malaysia3: IT4. Malaysia4: IT13. Malaysia5: IT3. Malaysia6: IT15. Indonesia1: D1-99, I187, I186. Indonesia2: SulA. Indonesia3: SulB. Vietnam1: IT16, IT17, IT18, IT20, IT21, IT22, IT23, IT25, IT27, IT28, IT29, IT30, IT31, IT32, IT33, IT34, IT35, IT36, IT37, IT39, IT40, IT41, IT42, IT43, IT45, IT46, IT47, IT48, IT49, IT50, V20, V16, V17, V19, V21, V26, V27, V28, V29, V30, V76, V100, V111, V112, V114, V157, V159, V160. Vietnam2: IT24. Vietnam3: V23. Honduras1: HE02. Honduras2: H-D2. Iran1: IR1, IR2, IR3, IR4, IR5, IR6, IR7. Iran2: IR8, IR9, IR10, IR11, IR12, IR13, IR14, IR15