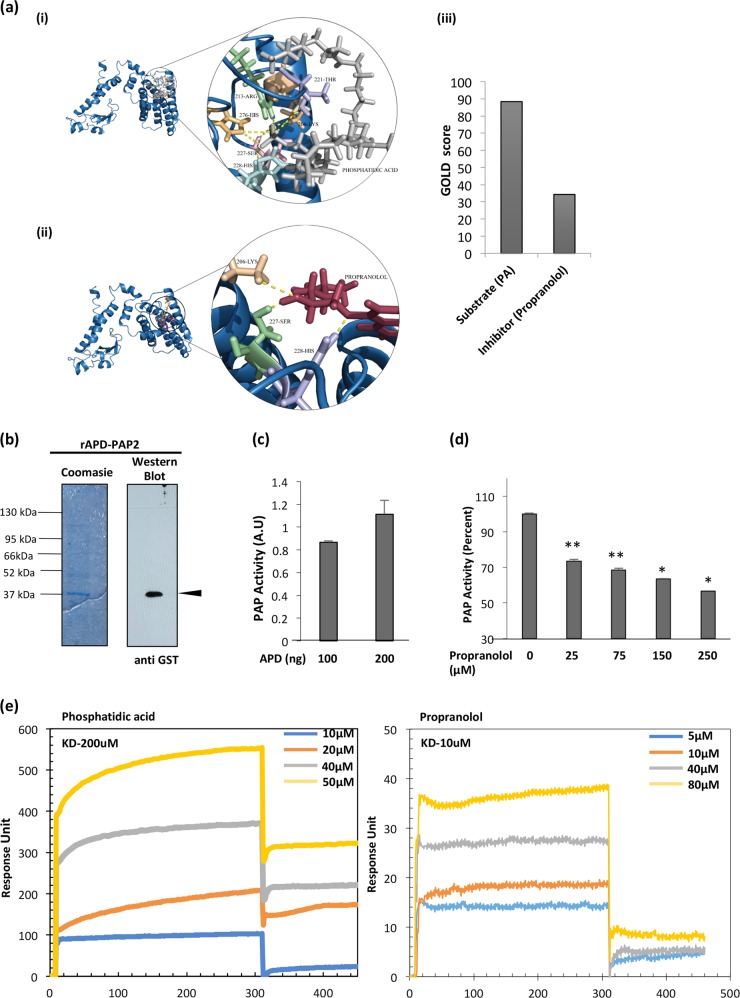
Fig. 2. Propranolol, a canonical inhibitor of PAP2 inhibits dephosphorylation of PA by PfPAP2.
a Active site prediction of PfPAP2 and docking with substrate PA and known PAP2 inhibitor, propranolol. Both ligands bind to the binding pocket with different affinity. (i) PA binds to the predicted active site that lies on APD with seven hydrogen bonds. (ii) Docking of known PAP2 inhibitor, propranolol against the active site present on APD (iii) GOLD score provides the quality of these docking represented in the form of bar graph. Substrate has a better score than inhibitor. b Expression of rAPD in the bacterial expression system. APD (~38.75 kDa) was eluted with reduced glutathione in Tris-NaCl buffer. Coomassie stained gels and corresponding Western blot analysis with anti-GST antibodies is shown confirming APD expression. A band corresponding to APD ~38.75 kDa was detected along with the degraded GST tag ~26 kDa in the eluted fractions. c Purified APD exhibited phosphatase activity. rAPD displayed phosphatase activity as suggested by malachite green assay. d Inhibition of APD activity by propranolol. Propranolol, its inhibitor, demonstrated dose-dependent inhibition of phosphatase activity of PfPAP2. *P < 0.05, **P < 0.01, (n = 3). e SPR analysis revealed binding of substrate, PA and inhibitor, propranolol to rAPD domain of PfPAP2. Dissociation kinetics of PA and propranolol was monitored with rAPD. PA demonstrated KD value of 200uM while propranolol demonstrated KD value of 10uM with rAPD domain