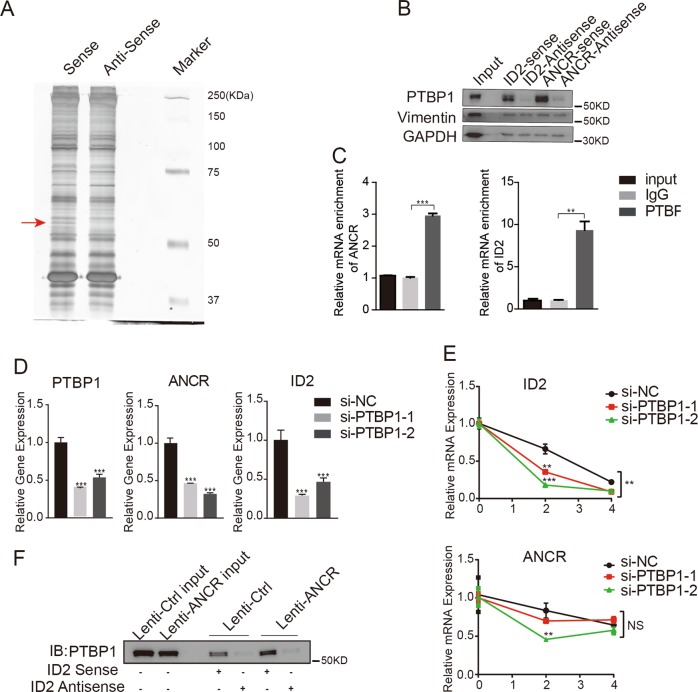
Fig. 7. ANCR regulates ID2 mRNA stability by binding to PTBP1.
a In vitro the RNA pull-down assay. hAMSCs lysates were incubated with biotin-labelled sense or antisense ANCR RNAs. After pull down, the proteins were subjected to SDS-PAGE and staining. The band indicated by the arrow was subjected to mass spectrometry. b Western blot analysis for PTBP1 or Vimentin following RNA pull down with biotin-labelled sense or antisense ID2 or ANCR. Antisense RNAs incubated with hAMSCs were used as nonspecific controls. c The RIP assay for PTBP1 enriched with ANCR (left) and ID2 (right) in hAMSCs. IgG was used as a negative control. All relative abundances were compared to 1% input. d PTBP1 was silenced in hAMSCs using two independent siRNAs (si-PTBP1-1 and si-PTBP1-2). The knockdown efficiency and expression of ANCR and ID2 were verified by qRT-PCR compared with si-NC. e hAMSCs transfected with si-PTBP1s or si-NC were treated with actinomycin D (5 μg/mL) and RNA was extracted at different time points (0, 2, and 4 h). The levels of ANCR and ID2 were analysed by qRT-PCR and normalized to GAPDH. mRNA at 0 h served as a reference. f The RNA pull down assay to determine the interaction between PTBP1 protein and ID2 mRNA in Lenti-Ctrl or Lenti-ANCR hAMSCs. Data are shown as the means ± S.D. (n = 3). *p < 0.05, **p < 0.01, and ***p < 0.001