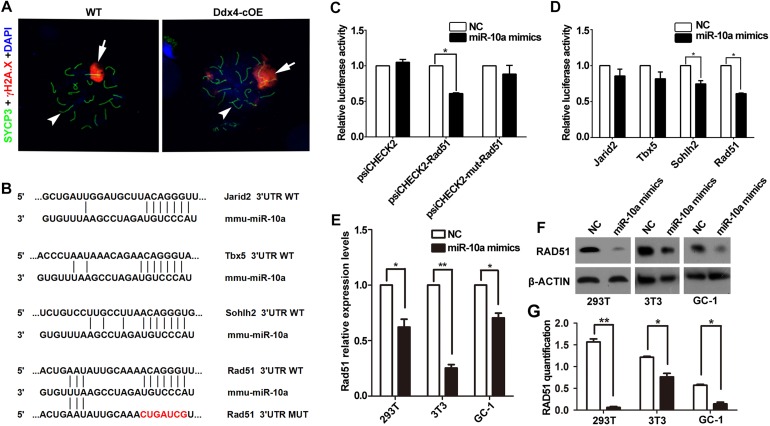
FIGURE 4.
miR-10a targets Rad51 is essential for DSB repair during meiosis. (A) Immunofluorescence staining of SYCP3 and γ-H2AX with DAPI at pachytene spermatocyte spreads in WT and Ddx4-cOE mice. Scale bars = 20 μm. Arrows indicate XY chromosome and arrowheads indicate autosomes. (B) A schematic representation of predicted miR-10a binding sites in the mouse Jarid2-3’UTR, Tbx5-3’UTR, Sohlh2-3’UTR and Rad51-3’UTR, respectively. (C) The relative luciferase activity was assayed following co-transfection of miR-10a mimics with the constructs encoding the wild-type or mutant miR-10a binding site of Rad51 3’-UTR into the HEK293T cells. The constructed reporter vectors (psiCHECK2 represent empty vector, psiCHECK2-Rad51 represent Rad51-3’UTR reporter plasmid and psiCHECK2-mut-Rad51 represent Rad51-mut-3’UTR reporter plasmid) are shown on the x-axis, and the y-axis represents the normalized luciferase activity (firefly luciferase activities were normalized to Renilla luciferase activities). The experiments were performed independently in triplicate. *P < 0.05. (D) The expression of luciferase Jarid2-3’UTR, Tbx5-3’UTR, Sohlh2-3’UTR and Rad51-3’UTR in HEK293T cells, respectively. The experiments were performed independently in triplicate. *P < 0.05. (E) RT-qPCR analyses of Rad51 expression level in miR-10a mimics co-transfected HEK293T, NIH3T3 and GC-1 cell lines, respectively. Data are presented as mean ± SEM (n = 3). *P < 0.05, ∗∗P < 0.01. (F) Protein level of RAD51 in miR-10a mimics co-transfected 293T, 3T3 and GC-1 cell lines by Western blot and protein quantification analyses, respectively. β-ACTIN as loading control. Protein quantification data are presented as mean ± SEM (n = 3). *P < 0.05, ∗∗P < 0.01. NC means negative control.