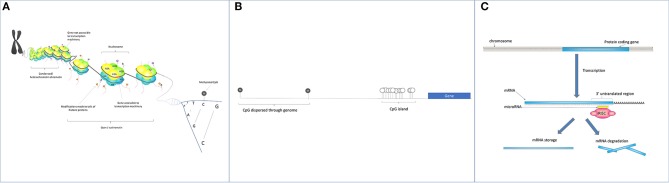
Figure 1.
(A) Histone modifications: The negative charge of DNA allows it to bind tightly to positively charged histone proteins. DNA wraps around octomers of histone proteins and forms discrete units known as nucleosomes, the basis of chromatin. The overall structure and openness of chromatin is dictated by chemical modifications of the N terminal amino acid tails of the histone proteins. Chemical modifications include acetylation, methylation, phosphorylation, SUMOylation, citrullination, and ADP-ribosylation. (B) CpG methylation: The majority of cytosines found in cytosine-guanine dinucleotides (gray circles) are methylated. CpG-rich sections of the genome (CpG islands) occurs in areas requiring transcriptional control e.g., retrotransposons and gene promotors. Here, methylation status is more dynamic, with some hypomethylated CpGs (white circles) facilitating promotor accessibility and gene transcription. (C) Small non-coding RNAs interact with complementary sequences in DNA and on mRNA to interfere with gene transcription and translation respectively. A well-known species of small RNAs are microRNAs. Mature single stranded microRNAs molecules (21–24 nt long) are incorporated into the RNA induced silencing complex (RISC) and then bind to a complementary sequence in the 3'UTRs of mRNA molecules. This binding inhibits mRNA translation and results in either mRNA degradation or storage.