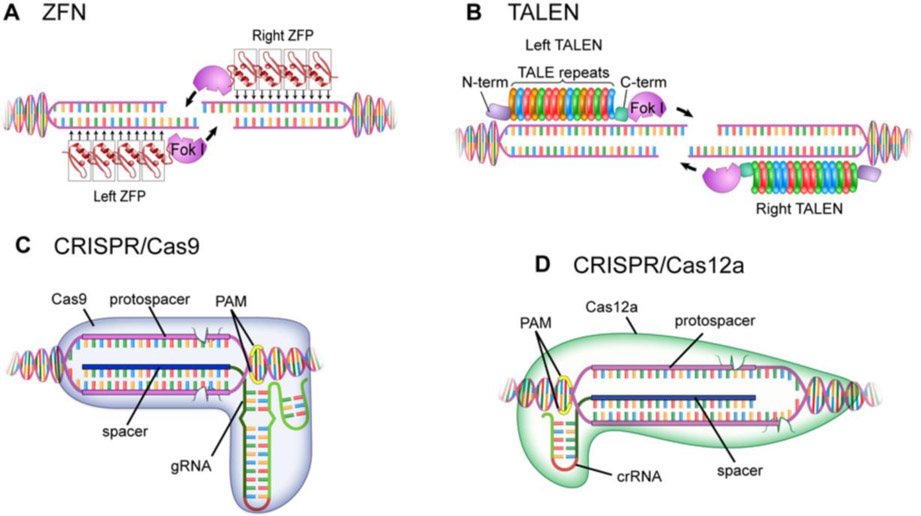
Figure 1. Commonly Used Programmable DNA Platforms:
A diagram showing programmable DNA binding platforms that recognize double-stranded DNA (dsDNA). A. A pair of 4-Finger Zinc Finger proteins binding to each side of the desired double-stranded break (DSB) location in the DNA. Each Zinc Finger (ZF) domain binds three bases of DNA; multiple ZF domains can be stringed together to bind longer stretches of DNA. When bound, the attached FokI nuclease (N) dimers become close enough in proximity to activate and catalyze a double stranded DNA break. B. A pair of Transcription Activator Like Effector domains (TALEs) bound to each side of the preferred DSB position. TALE domains consist of a series of 35 amino acid repeats attached in sequence. Each of these motifs binds to a single specific DNA base and can be strung together to recognize diverse DNA sequences. C. The CRISPR/Cas9 endonuclease system functions through the interaction of a RNA guide with a single protein, Cas9. The RNA guide consists of two domains, a constant poly-hairpin structure that interacts with the Cas9 protein and a programmable guide region that targets DNA through standard Watson-Crick base pairing. Upon binding its target region by interrogating and unwinding (melting) the dsDNA, the Cas9 protein induces a blunt double stranded break. D. Another CRISPR system makes use of different class of guide RNA coupled with a different constant protein, Cas12a. Targeting is again determined by Watson-Crick base pairing between the guide RNA and the DNA, following which Cas12a induces a DSB with its signature overhang.