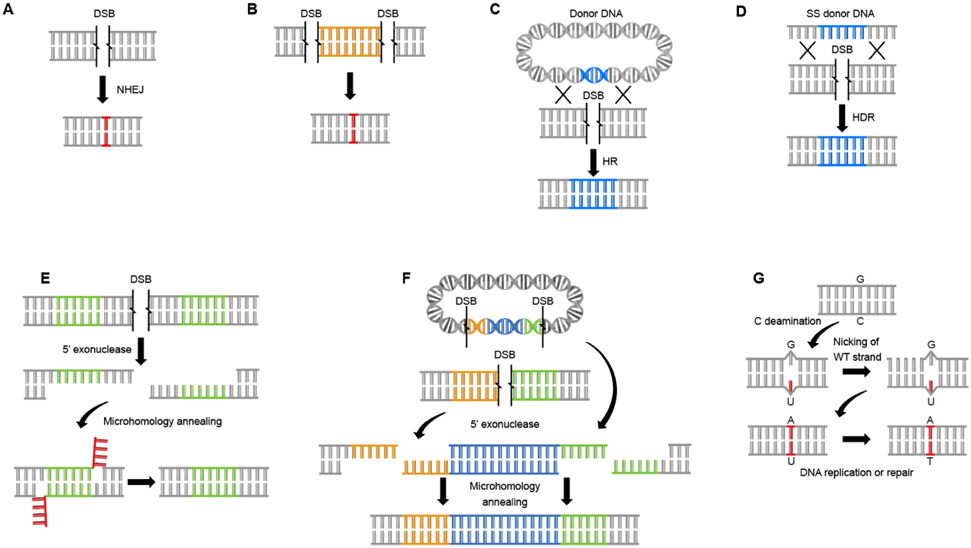
Figure 2. Associated Biochemical Activities Critical For Precision Gene Editing:
Diagram showing the different cellular repair machineries and corresponding biochemical functions critical for gene editing. A. Repair of a DSB by Non-Homologous End Joining (NHEJ) introduces mutation through the creation of small insertions or deletions (indels). B. NHEJ repair can delete long segments of DNA (whole genes) by creating two double stranded breaks and removing the intervening region from the chromosome C. Homologous Recombination (HR) can be used to insert exogenous DNA though the introduction of an exogenous template flanked by large (often >500 base pairs) dsDNA sequences homologous the sequence adjacent to the DSB. D. Oligo-directed HDR can introduce small changes through the introduction of a short single-strand DNA (ssDNA) template flanked with homology matching the regions to either side of the DSB. E. Micro-Homology Mediated End Joining can be used to create small reproducible deletions. This repair pathway functions by annealing small homologous regions on each side of the DSB. Unbound DNA flaps are removed and ends are ligated resulting in the removal of one homology arm and the intervening region. F. MMEJ can also be used to insert exogenous DNA through the introduction of a template with matching microhomology arms. G. Single nucleotide polymorphisms can be introduced without a DSB using a base editor. By targeting a cytosine deaminase to a specific target in the genome it is possible to convert a cytosine to a uracil resulting in a mismatch base. This mismatch is then recognized and repaired, generating a single base change depending on which strand is chosen as repair template, this can be selected by design by repeated nicking of the strand to be modified.