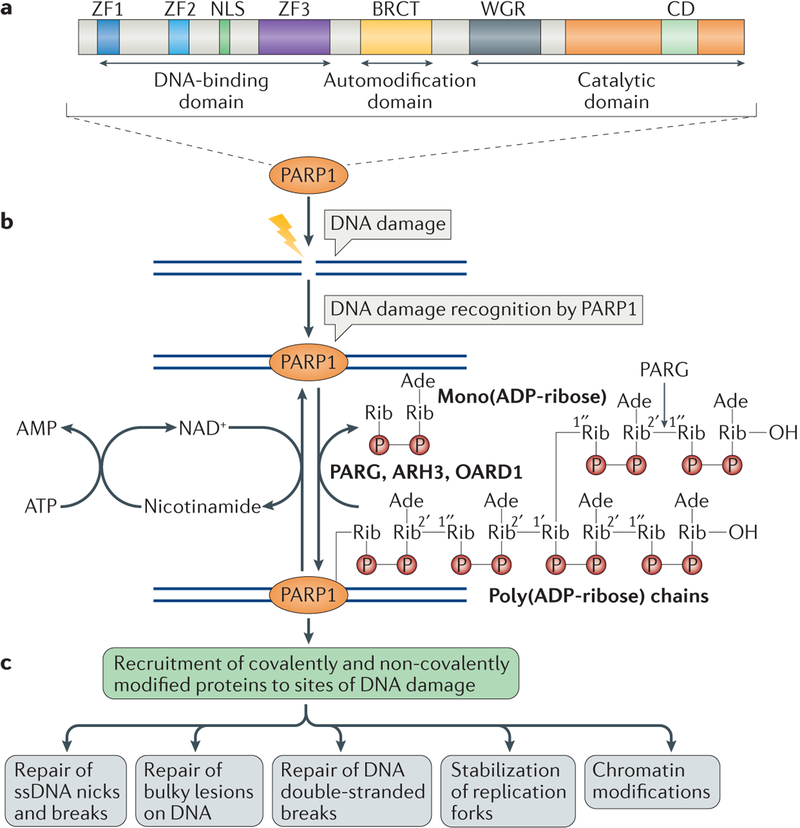
Figure 1 |. The biochemical functions of poly(ADP-ribose) polymerase 1 in DNA damage repair.
a | The domains of poly(ADP-ribose) polymerase 1 (PARP1) are shown. PARP1 has a DNA-binding domain (DBD), which consists of three zinc finger motifs (ZF1–3) and also contains a nuclear localization signal (NLS); a central automodification domain, which contains an interaction motif termed the BRCA1 C terminus (BRCT) domain; and a carboxy-terminal catalytic domain that contains a signature of PARP proteins, the conserved domain (CD). The CD contains the active site and binds to NAD+ and also to the Trp-Gly-Arg (WGR) domain. b | PARP1 detects DNA damage through its DBD, and it is activated by synthesizing poly(ADP-ribose) (PAR) chains mainly on itself but also on some of its target proteins. NAD+ is used as substrate for PAR formation, and is replenished in the cells from nicotinamide, using ATP. PAR chains are rapidly catabolized by PAR glycohydrolase (PARG), ADP-ribosylhydrolase 3 (ARH3) and O-acyl-ADP-ribose deacylase 1 (OARD1). PARG cleaves the 2′,1″-O-glycosidic ribose–ribose bonds of PAR. However, cleavage of the terminal ADP-ribose moiety requires OARD1 and results in the release of mono(ADP-ribose). c | Poly(ADP)ribosylation (PARylation) of PARP1 and other target proteins, both covalently and non-covalently, results in the recruitment of multiple proteins that have roles in different aspects of DNA damage repair. Ade, adenosine; P, phosphate residue; Rib, ribose moiety; ssDNA, single-stranded DNA.