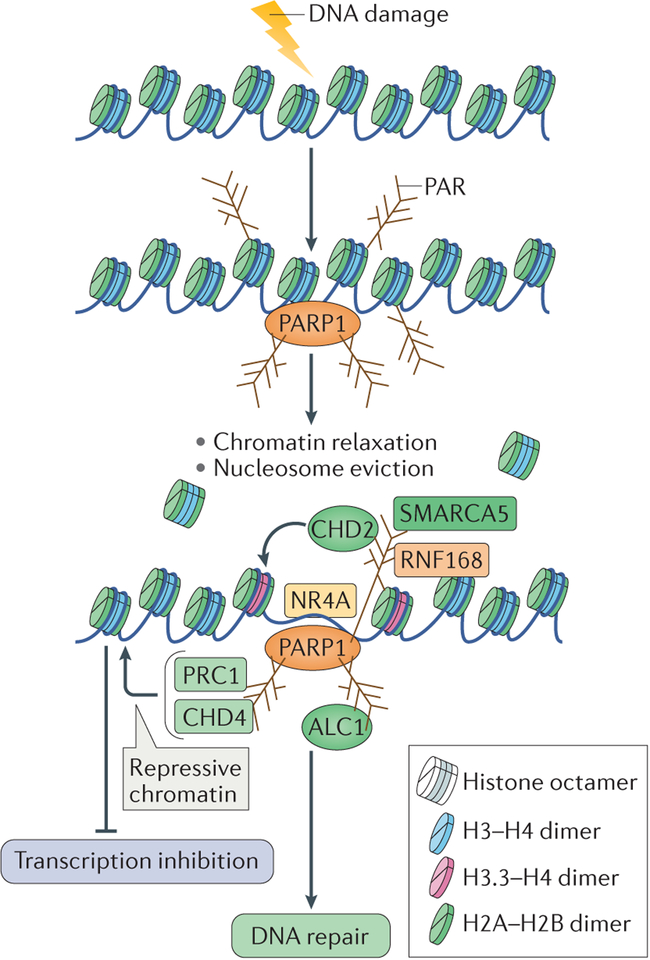
Figure 5 |. Chromatin changes induced by poly(ADP-ribose) polymerase 1 — integrating DNA repair.
DNA damage in the context of chromatin results in poly(ADP-ribose) polymerase 1 (PARP1) activation and autopoly(ADP)ribosylation (autoPARylation). PARP1 also PARylates histone tails, which results in chromatin relaxation and in nucleosome eviction from the DNA. This also allows the recruitment of several chromatin remodellers through their binding to PAR, which further relax chromatin to facilitate DNA repair. These chromatin remodellers include amplified in liver cancer protein 1 (ALC1), SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (SMARCA5), chromodomain helicase DNA-binding protein 2 (CHD2), which also incorporates the histone variant H3.3 into nucleosomes, and the transcription factor NR4A (a member of the nuclear orphan receptors). Conversely, repressive chromatin modifiers, such as CHD4 and Polycomb repressive complex 1 (PRC1), are also recruited to sites of DNA damage by binding to PAR. They might be recruited to inhibit transcription at flanking regions to facilitate DNA repair. RNF168, RING finger protein 168.