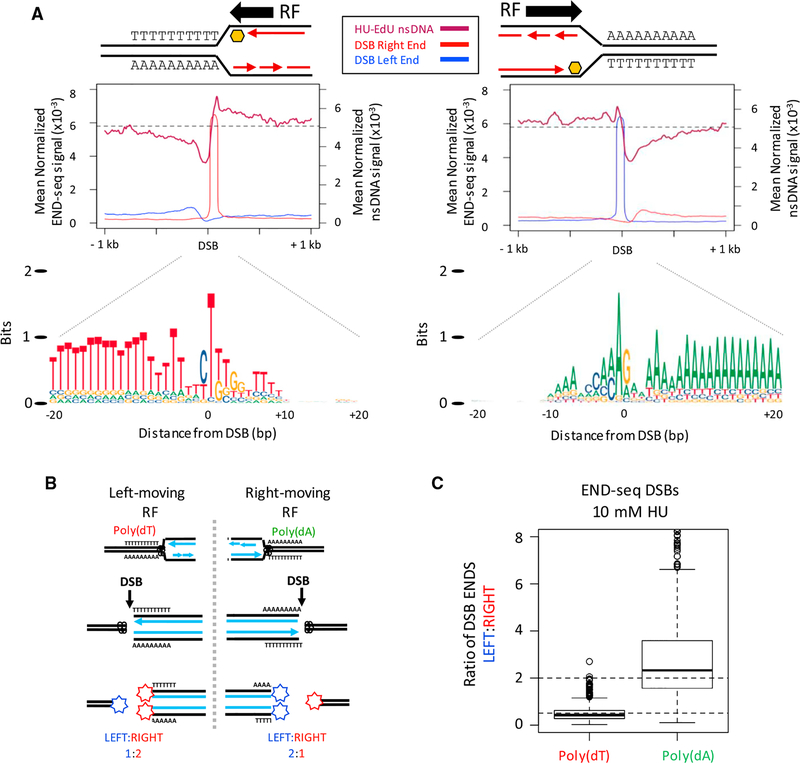
Figure 4. Unidirectional Replication Fork Stalling and Breakage Are Associated with poly(dA:dT).
(A) (Top panels) Composite plots for nsDNA and strand-separated END-seq reads (DSB left/right end) at the top-1,000 poly(dA) or poly(dT) DSB sites in activated B cells treated with 10 mM HU. Replication fork (RF) direction is inferred from the nsDNA signal surrounding the DSB. (Bottom panel) Composite motif ± 20 bp of the sharp DSB end for the regions analyzed in top panels, illustrating a unidirectional RF stall at poly(dA:dT) containing poly(dT) on the leading strand.
(B) Proposed ratios of DSB ends at a poly(dA:dT) polar replication fork barrier.
(C) Boxplots showing the ratio of DSB ends (left:right) for the top-1,000 DSB sites with poly(dA) or poly(dT) near the sharp DSB end in activated B cells treated with 10 mM HU.