Abstract
Background
The comparison of expression QTL (eQTL) maps obtained in different tissues is an essential step to understand how gene expression is genetically regulated in a context-dependent manner. In the current work, we have compared the transcriptomic and eQTL profiles of two porcine tissues (skeletal muscle and liver) which typically show highly divergent expression profiles, in 103 Duroc pigs genotyped with the Porcine SNP60 BeadChip (Illumina) and with available microarray-based measurements of hepatic and muscle mRNA levels. Since structural variation could have effects on gene expression, we have also investigated the co-localization of cis-eQTLs with copy number variant regions (CNVR) segregating in this Duroc population.
Results
The analysis of differential expresssion revealed the existence of 1204 and 1490 probes that were overexpressed and underexpressed in the gluteus medius muscle when compared to liver, respectively (|fold-change| > 1.5, q-value < 0.05). By performing genome scans in 103 Duroc pigs with available expression and genotypic data, we identified 76 and 28 genome-wide significant cis-eQTLs regulating gene expression in the gluteus medius muscle and liver, respectively. Twelve of these cis-eQTLs were shared by both tissues (i.e. 42.8% of the cis-eQTLs identified in the liver were replicated in the gluteus medius muscle). These results are consistent with previous studies performed in humans, where 50% of eQTLs were shared across tissues. Moreover, we have identified 41 CNVRs in a set of 350 pigs from the same Duroc population, which had been genotyped with the Porcine SNP60 BeadChip by using the PennCNV and GADA softwares, but only a small proportion of these CNVRs co-localized with the cis-eQTL signals.
Conclusion
Despite the fact that there are considerable differences in the gene expression patterns of the porcine liver and skeletal muscle, we have identified a substantial proportion of common cis-eQTLs regulating gene expression in both tissues. Several of these cis-eQTLs influence the mRNA levels of genes with important roles in meat (CTSF) and carcass quality (TAPT1), lipid metabolism (TMEM97) and obesity (MARC2), thus evidencing the practical importance of dissecting the genetic mechanisms involved in their expression.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5889-5) contains supplementary material, which is available to authorized users.
Background
The performance of GWAS in humans has revealed that most of the regions that display significant associations with complex traits are not exonic, meaning that regulatory polymorphisms might have important effects on phenotypic variation [1]. This realization has prompted the mapping of expression QTL (eQTL), i.e. single nucleotide polymorphisms (SNP), indels or copy number variants (CNV) that explain part of the variance of gene expression phenotypes [1]. Such studies have revealed that the majority of eQTL exert their effects in cis- (i.e. on neighboring genes) [2]. There are also evidences that CNV-tagging SNPs are enriched in cis-eQTLs and that they often modulate multiple expression traits [3]. By examining the patterns of expression of 22,286 genes in 9 human tissues, the GTEx Consortium has shown that approximately 50% of eQTL are shared by the nine tissues and that most of them display consistent effects across tissues [4].
In pigs, hundreds of eQTLs with effects on muscle [5–9], liver [10, 11] and backfat [12] gene expression have been mapped. Often, these pig eQTL studies have targeted subsets of genes either mapping to QTL [12, 13] or displaying significant expression-phenotype correlations [5, 6, 10, 11]. The broad majority of porcine eQTL studies have targeted single anatomic locations and, in consequence, they do not provide clues about the differential genetic regulation of distinct tissues and organs. Moreover, these genome scans have explored the association of gene expression with the allelic variation of SNPs or microsatellites, neglecting the potential effect of CNVs on expression phenotypes. The goals of the current work were to compare the gene expression patterns and cis-eQTL landscapes of the pig gluteus medius (GM) skeletal muscle and liver, two tissues with highly differentiated patterns of expression [13], as well as to investigate the co-localization of cis-eQTLs and CNVRs.
Results
Differential expression analysis
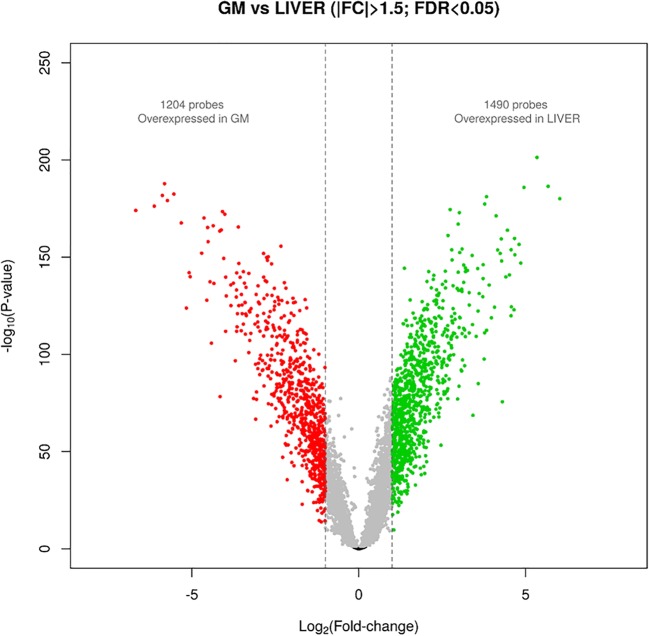
A total of 1204 probes were found to be overexpressed in the GM muscle (|FC| > 1.5; q-value < 0.05), whereas 1490 showed overexpression in the liver (Fig. 1 and Additional file 1). The list of genes showing the highest levels of differential expression between muscle and liver, included the bridging integrator 1 (BIN1, log2FC = 5.34, q-value = 2.21E-198), the sorbitol dehydrogenase (SORD, log2FC = − 5.82, q-value = 3.47E-185), the protein phosphatase 1 regulatory inhibitor subunit 1A (PPP1R1A, log2FC = 5.67, q-value = 5.62E-184), the zinc finger protein 106 (ZNF106, log2FC = 4.94, q-value = 1.60E-183), the hydroxysteroid 17-beta dehydrogenase 13 (HSD17B13, log2FC = − 5.54, q-value = 3.04E-180) and apolipoprotein A1 (APOA1, log2FC = − 5.88, q-value = 1.25E-179). Noteworthy, the log2FC values are very high and the q-values highly significant, thereby evidencing that the expression profiles of the porcine liver and the skeletal muscle are strongly divergent.
Fig. 1.
Volcano plot depicting porcine genes that are differentially expressed in the porcine gluteus medius muscle and the liver
Detection of expression QTL in the porcine skeletal muscle and liver
A total of 76 and 28 genome-wide significant cis-eQTLs were identified in the GM muscle and liver, respectively (Tables 1 and 2). Several genes that are cis-regulated in the muscle participate in lipid or carbohydrate metabolism, e.g. HIV-1 Tat Interactive Protein 2 (HTATIP2) [14], exocyst complex component 7 (EXOC7) [15] acyl-CoA dehydrogenase short chain (ACADS) [16], insulin degrading enzyme (IDE) [17], solute carrier family 38 member 9 (SLC38A9) [18], and family with sequence similarity 3 member C (FAM3C) [19]. With regard to the liver, hydroxysteroid dehydrogenase like 2 (HSDL2) [20] and lipase A (LIPA) [21] genes have also important roles in lipid metabolism. The level of overlap between the muscle and the liver cis-eQTL data sets was considerable, with 12 cis-eQTLs shared by both tissues (42.8% of the cis-eQTLs detected in the liver were also detected in the GM muscle). Four examples of shared cis-eQTLs are shown in Fig. 2. There was a strong consistency in the direction of the effects of the shared cis-eQTLs in both tissues. For instance, the cis-eQTL regulating ARL2BP and TMEM97 showed positive allelic effects in both tissues, whilst consistent negative allelic effects were estimated for the cis-eQTL modulating the hepatic and muscular expression of BSDC1, POLR3D and TAP1 (Tables 1 and 2). We also detected a number of cis-eQTLs that had effects on gene expression either in the muscle or in the liver (Figs. 3 and 4). RNA-Seq data from the GM muscle from 52 of the 103 pigs with GM muscle microarray was also available. Comparison of the genotypic means obtained with both platforms and corresponding to genes that are cis-regulated in the GM muscle provided highly consistent results, i.e. the direction of the difference between the two homozygous genotypes was concordant in 82% of the comparisons and the same ordering of genotypes (from highest to lowest expression) was obtained in 70% of the comparisons (Additional file 2). However, both methods did not always yield significant results (about 50% of coincidence with regard to statistical significance). The most probable reason for this is that the sample size of the RNA-Seq data set is half the size when compared to the microarray data set, which leads to larger standard deviations for the RNA-Seq datasets. This would consequently reduce the power to detect significant differences between the genotypic means.
Table 1.
List of the genome-wide significant cis-eQTLs detected in the gluteus medius (GM) muscle (those that have been consistently detected in the liver are shown in bold)1
| Muscle cis-eQTL | Gene | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSC | N | SNP | Region (Mb) | P-value | q-value | B | δ ± SE | A1 | MAF | Acronym | Region (Mb) |
| 1 | 51 | ASGA0000817 | 0.2–9.9 | 6.69E-08 | 2.12E-04 | 1.91E-03 | −0.37 ± 0.06 | A | 0.388 | ARID1B | 9.7–10.2 |
| 1 | 15 | ALGA0002427 | 32.2–36.6 | 2.74E-11 | 5.90E-07 | 7.80E-07 | 0.58 ± 0.06 | A | 0.365 | AKAP7 | 32.2–32.3 |
| 1 | 10 | INRA0003613 | 101.9–108.9 | 3.29E-12 | 2.00E-08 | 9.00E-08 | −0.29 ± 0.03 | C | 0.433 | WDR7 | 106.0–106.4 |
| 1 | 2 | MARC0039560 | 125.9–128.7 | 5.67E-06 | 3.24E-02 | 1.62E-01 | −0.16 ± 0.03 | G | 0.481 | HYPK | 127.7–127.7 |
| 1 | 5 | DIAS0000210 | 183.3–183.9 | 5.36E-07 | 7.63E-03 | 1.53E-02 | 0.28 ± 0.05 | C | 0.466 | CNIH1 | 183.8–183.8 |
| 1 | 9 | ASGA0006202 | 231.0–239.7 | 3.49E-11 | 8.10E-07 | 9.90E-07 | 0.50 ± 0.07 | A | 0.115 | GRHPR | 238.1–238.1 |
| 1 | 18 | M1GA0001375 | 240.2–249.6 | 1.92E-13 | 1.00E-08 | 1.00E-08 | 0.27 ± 0.03 | A | 0.399 | ALG2 | 240.9–240.9 |
| 1 | 1 | H3GA0004649 | 260.9–260.9 | 2.36E-06 | 3.24E-02 | 6.73E-02 | −0.36 ± 0.07 | A | 0.17 | PSMD5 | 260.8–260.8 |
| 2 | 1 | ALGA0011482 | 5.8–5.8 | 1.47E-06 | 4.20E-02 | 4.20E-02 | 0.28 ± 0.06 | A | 0.144 | CTSF | 5.8–5.8 |
| 2 | 22 | MARC0095742 | 32.5–39.3 | 4.52E-10 | 4.30E-06 | 1.29E-05 | −0.53 ± 0.08 | A | 0.354 | HTATIP2 | 39.0–39.0 |
| 2 | 24 | ALGA0013197 | 33.0–39.3 | 1.24E-14 | 0.00E+ 00 | 0.00E+ 00 | −0.52 ± 0.06 | A | 0.25 | PRMT3 | 38.8–39.0 |
| 2 | 4 | ALGA0115180 | 115.1–116.1 | 9.98E-09 | 2.36E-04 | 2.84E-04 | 0.37 ± 0.06 | A | 0.141 | WDR36 | 115.8–115.9 |
| 2 | 8 | DIAS0000550 | 140.2–145.0 | 1.54E-06 | 1.97E-02 | 4.40E-02 | −0.31 ± 0.05 | G | 0.466 | ETF1 | 140.4–140.5 |
| 2 | 15 | H3GA0055810 | 147.1–148.7 | 2.30E-08 | 6.56E-04 | 6.56E-04 | −0.30 ± 0.05 | G | 0.16 | LARS | 147.4–147.5 |
| 3 | 1 | MARC0045458 | 3.6–3.6 | 3.41E-08 | 9.72E-04 | 9.72E-04 | 0.26 ± 0.04 | G | 0.223 | AP5Z1 | 3.6–3.6 |
| 3 | 2 | MARC0020406 | 4.1–4.2 | 4.17E-08 | 7.28E-04 | 1.19E-03 | −0.68 ± 0.12 | A | 0.121 | ZNF12 | 4.4–4.4 |
| 3 | 14 | ALGA0017913 | 18.6–19.8 | 1.33E-09 | 1.78E-05 | 3.78E-05 | 0.30 ± 0.05 | G | 0.125 | NSMCE1 | 19.6–19.6 |
| 3 | 2 | DRGA0003838 | 29.6–29.6 | 5.05E-07 | 7.81E-03 | 1.44E-02 | −0.37 ± 0.06 | A | 0.37 | MKL2 | 28.9–29.2 |
| 3 | 26 | MARC0101576 | 30.0–34.4 | 5.74E-09 | 1.64E-04 | 1.64E-04 | −0.22 ± 0.04 | G | 0.345 | RSL1D1 | 31.2–31.2 |
| 3 | 17 | MARC0000159 | 48.6–54.6 | 2.77E-06 | 9.87E-03 | 7.89E-02 | −0.27 ± 0.06 | G | 0.194 | MRPS9 | 49.6–49.6 |
| 3 | 18 | MARC0049447 | 93.7–97.2 | 3.90E-07 | 2.78E-03 | 1.11E-02 | 0.29 ± 0.05 | A | 0.272 | THADA | 96.8–97.1 |
| 3 | 7 | M1GA0026608 | 126.9–127.8 | 4.55E-09 | 6.48E-05 | 1.30E-04 | 0.58 ± 0.10 | A | 0.178 | CYS1 | 126.3–126.4 |
| 4 | 6 | ASGA0018519 | 14.3–15.8 | 3.76E-06 | 3.03E-02 | 1.07E-01 | −0.21 ± 0.04 | C | 0.393 | NDUFB9 | 15.1–15.1 |
| 4 | 2 | ASGA0018539 | 14.6–14.6 | 9.98E-09 | 2.84E-04 | 2.84E-04 | 0.19 ± 0.03 | G | 0.322 | WASHC5 | 14.5–14.6 |
| 4 | 6 | ALGA0024302 | 30.2–31.5 | 2.51E-06 | 9.62E-03 | 7.15E-02 | −0.40 ± 0.07 | G | 0.433 | OXR1 | 30.7–31.2 |
| 4 | 11 | DIAS0000176 | 95.2–99.8 | 2.07E-07 | 1.97E-03 | 5.91E-03 | 0.33 ± 0.06 | A | 0.252 | APH1A | 98.8–98.8 |
| 4 | 4 | M1GA0006152 | 95.3–96.1 | 3.55E-07 | 1.01E-02 | 1.01E-02 | −0.23 ± 0.04 | G | 0.341 | S100A13 | 96.0–96.0 |
| 5 | 4 | ASGA0104541 | 20.3–23.5 | 1.61E-06 | 1.57E-02 | 4.59E-02 | 0.17 ± 0.03 | A | 0.325 | TSPAN31 | 23.0–23.0 |
| 5 | 12 | SIRI0000318 | 25.6–29.4 | 2.96E-07 | 2.11E-03 | 8.43E-03 | −0.53 ± 0.10 | A | 0.306 | TMEM5 | 28.2–28.2 |
| 5 | 24 | MARC0113779 | 80.3–84.7 | 2.66E-10 | 7.10E-07 | 7.58E-06 | −0.39 ± 0.06 | A | 0.136 | C12orf73 | 80.5–80.5 |
| 6 | 25 | ASGA0098408 | 17.0–21.7 | 1.11E-08 | 1.40E-04 | 3.16E-04 | 0.49 ± 0.08 | G | 0.24 | ARL2BP | 19.2–19.2 |
| 6 | 28 | DIAS0004785 | 45.5–55.4 | 4.23E-06 | 1.72E-02 | 1.21E-01 | −0.15 ± 0.03 | A | 0.471 | RABAC1 | 49.9–49.9 |
| 6 | 13 | ASGA0094600 | 70.1–71.6 | 8.61E-07 | 7.64E-03 | 2.45E-02 | −0.33 ± 0.06 | G | 0.296 | LZIC | 70.2–70.2 |
| 6 | 17 | MARC0031169 | 70.1–71.6 | 3.65E-06 | 1.16E-02 | 1.04E-01 | −0.19 ± 0.03 | G | 0.279 | DFFA | 70.7–70.7 |
| 6 | 36 | ASGA0028827 | 81.9–89.9 | 5.30E-12 | 1.00E-08 | 1.50E-07 | −0.39 ± 0.05 | G | 0.238 | BSDC1 | 88.8–88.8 |
| 6 | 52 | MARC0015713 | 90.6–99.9 | 2.96E-08 | 4.21E-04 | 8.43E-04 | −0.20 ± 0.03 | A | 0.379 | AFG3L2 | 97.2–97.2 |
| 6 | 2 | ALGA0122704 | 95.9–99.4 | 3.18E-06 | 6.66E-03 | 9.05E-02 | 0.37 ± 0.08 | A | 0.327 | RAB12 | 99.2–99.2 |
| 6 | 6 | ALGA0037549 | 157.5–158.8 | 2.07E-08 | 5.89E-04 | 5.89E-04 | 0.29 ± 0.05 | A | 0.262 | MRPL37 | 158.0–158.1 |
| 6 | 1 | ASGA0030033 | 158.6–158.6 | 5.27E-06 | 2.15E-02 | 1.50E-01 | −0.22 ± 0.05 | G | 0.359 | YIPF1 | 158.4–158.4 |
| 7 | 19 | ASGA0035147 | 86.3–89.1 | 1.55E-12 | 4.00E-08 | 4.00E-08 | 0.41 ± 0.05 | A | 0.5 | MTHFD1 | 88.4–88.5 |
| 7 | 21 | ALGA0043682 | 92.1–99.8 | 7.12E-09 | 2.03E-04 | 2.03E-04 | −0.26 ± 0.04 | G | 0.333 | DCAF5 | 92.7–92.8 |
| 8 | 11 | ALGA0112294 | 10.6–17.6 | 1.63E-09 | 6.63E-06 | 4.64E-05 | −0.25 ± 0.04 | C | 0.399 | TAPT1 | 11.3–11.4 |
| 8 | 6 | ALGA0049334 | 117.4–118.2 | 9.95E-09 | 8.85E-05 | 2.84E-04 | −0.29 ± 0.05 | A | 0.335 | BDH2 | 117.9–117.9 |
| 9 | 37 | ASGA0042057 | 20.2–24.2 | 2.61E-10 | 7.44E-06 | 7.44E-06 | 0.56 ± 0.08 | G | 0.49 | CTSC | 21.7–21.7 |
| 9 | 9 | ASGA0043022 | 40.3–47.6 | 8.95E-11 | 2.55E-06 | 2.55E-06 | −0.28 ± 0.03 | A | 0.165 | ARHGEF12 | 47.4–47.6 |
| 10 | 4 | ASGA0046443 | 10.0–14.1 | 1.10E-16 | 0.00E+ 00 | 0.00E+ 00 | −0.92 ± 0.09 | G | 0.228 | MARC2 | 10.1–10.1 |
| 10 | 11 | ASGA0048312 | 51.7–58.7 | 5.83E-08 | 1.66E-03 | 1.66E-03 | −0.33 ± 0.04 | A | 0.409 | COMMD3 | 52.5–52.6 |
| 10 | 2 | ALGA0059373 | 54.1–54.4 | 4.39E-08 | 6.26E-04 | 1.25E-03 | 0.33 ± 0.06 | G | 0.298 | PLXDC2 | 54.2–54.6 |
| 11 | 3 | DIAS0002284 | 20.7–21.0 | 3.50E-08 | 9.98E-04 | 9.98E-04 | −0.21 ± 0.04 | A | 0.296 | ESD | 20.6–20.6 |
| 12 | 4 | ALGA0115312 | 4.2–4.8 | 4.24E-07 | 1.21E-02 | 1.21E-02 | −0.20 ± 0.04 | G | 0.296 | ST6GALNAC2 | 4.9–4.9 |
| 12 | 3 | DRGA0011563 | 4.2–4.8 | 3.95E-06 | 3.75E-02 | 1.13E-01 | −0.42 ± 0.07 | A | 0.17 | EXOC7 | 5.3–5.3 |
| 12 | 3 | ALGA0064915 | 10.9–11.6 | 1.19E-06 | 1.70E-02 | 3.39E-02 | −0.40 ± 0.08 | G | 0.291 | ABCA5 | 11.0–11.1 |
| 12 | 16 | MARC0016809 | 20.3–24.8 | 3.26E-12 | 9.00E-08 | 9.00E-08 | 0.49 ± 0.06 | G | 0.272 | PNPO | 24.1–24.2 |
| 12 | 2 | DIAS0000973 | 23.5–23.6 | 1.52E-06 | 2.34E-02 | 4.33E-02 | 0.24 ± 0.04 | C | 0.257 | MRPL10 | 24.0–24.0 |
| 12 | 8 | DIAS0000242 | 40.0–48.8 | 3.14E-06 | 2.01E-02 | 8.96E-02 | 0.25 ± 0.05 | G | 0.433 | TMEM98 | 42.0–42.1 |
| 12 | 6 | ASGA0054801 | 44.3–46.5 | 1.28E-06 | 2.18E-02 | 3.66E-02 | 0.20 ± 0.04 | G | 0.481 | TMEM97 | 44.6–44.6 |
| 12 | 7 | ALGA0066917 | 50.5–59.3 | 4.57E-11 | 4.50E-07 | 1.30E-06 | 0.37 ± 0.05 | G | 0.335 | RABEP1 | 51.6–51.8 |
| 12 | 1 | ALGA0120796 | 51.9–51.9 | 2.27E-07 | 6.46E-03 | 6.46E-03 | −0.13 ± 0.02 | C | 0.412 | PSMB6 | 52.0–52.0 |
| 12 | 7 | MARC0002558 | 59.0–59.9 | 3.63E-10 | 1.03E-05 | 1.03E-05 | 0.41 ± 0.05 | A | 0.35 | ALDH3A2 | 59.9–59.9 |
| 13 | 2 | ALGA0067450 | 2.5–2.7 | 7.69E-07 | 2.19E-02 | 2.19E-02 | 0.34 ± 0.06 | A | 0.306 | SH3BP5 | 2.5–2.5 |
| 13 | 9 | ASGA0086643 | 12.4–14.4 | 1.64E-12 | 1.00E-08 | 5.00E-08 | 0.52 ± 0.06 | A | 0.422 | NGLY1 | 12.6–12.6 |
| 13 | 5 | ALGA0068741 | 19.4–23.1 | 1.67E-06 | 2.38E-02 | 4.77E-02 | −0.16 ± 0.03 | G | 0.269 | PDCD6IP | 19.5–19.6 |
| 13 | 5 | MARC0000523 | 31.1–32.5 | 3.32E-11 | 9.50E-07 | 9.50E-07 | −0.35 ± 0.05 | A | 0.155 | MAPKAPK3 | 33.0–33.1 |
| 14 | 12 | H3GA0038597 | 6.1–7.2 | 6.65E-13 | 2.00E-08 | 2.00E-08 | −0.23 ± 0.03 | A | 0.383 | POLR3D | 6.5–6.5 |
| 14 | 1 | SIRI0001291 | 22.7–22.7 | 4.88E-08 | 1.39E-03 | 1.39E-03 | −0.20 ± 0.03 | G | 0.291 | PGAM5 | 22.7–22.7 |
| 14 | 8 | MARC0094155 | 40.6–43.2 | 4.74E-10 | 5.71E-06 | 1.35E-05 | −0.44 ± 0.06 | G | 0.212 | ACADS | 40.6–40.6 |
| 14 | 5 | ASGA0065742 | 102.7–106.9 | 3.84E-08 | 5.48E-04 | 1.10E-03 | −0.34 ± 0.06 | A | 0.147 | IDE | 103.9–104.1 |
| 14 | 4 | MARC0043866 | 110.5–111.0 | 2.01E-06 | 2.86E-02 | 5.72E-02 | 0.30 ± 0.05 | A | 0.49 | COX15 | 110.8–110.8 |
| 14 | 11 | ALGA0081813 | 124.1–125.1 | 1.19E-08 | 1.70E-04 | 3.39E-04 | 0.44 ± 0.07 | G | 0.277 | CASP7 | 124.0–124.0 |
| 15 | 11 | ASGA0069526 | 42.2–49.0 | 7.12E-13 | 0.00E+ 00 | 2.00E-08 | 1.33 ± 0.16 | G | 0.067 | SORBS2 | 46.5–46.9 |
| 15 | 54 | ALGA0085678 | 54.5–69.7 | 4.49E-08 | 1.28E-03 | 1.28E-03 | −0.36 ± 0.06 | G | 0.112 | UGGT1 | 59.1–59.2 |
| 15 | 9 | ASGA0070490 | 116.4–118.5 | 1.07E-10 | 3.05E-06 | 3.05E-06 | 0.35 ± 0.05 | A | 0.471 | XRCC5 | 118.3–118.4 |
| 16 | 1 | ALGA0090252 | 34.2–34.2 | 1.21E-06 | 3.45E-02 | 3.45E-02 | −0.24 ± 0.04 | C | 0.485 | SLC38A9 | 34.7–34.8 |
| 16 | 6 | MARC0049616 | 72.0–73.1 | 7.13E-08 | 1.46E-03 | 2.03E-03 | −0.32 ± 0.05 | A | 0.288 | CCT5 | 72.2–72.2 |
| 17 | 2 | ALGA0096195 | 57.0–57.1 | 9.93E-08 | 1.42E-03 | 2.83E-03 | −0.36 ± 0.05 | A | 0.223 | FAM210B | 56.9–56.9 |
| 18 | 5 | ALGA0097540 | 24.4–25.8 | 1.38E-07 | 3.94E-03 | 3.94E-03 | 0.44 ± 0.08 | A | 0.142 | FAM3C | 25.5–25.6 |
1SSC: porcine chromosome, N: Number of SNPs significantly associated with the trait under study, SNP: SNP displaying the most significant association with the trait under study, Region (Mb): region containing SNPs significantly associated with the trait under study, P-value: nominal P-value, q-value: q-value calculated with a false discovery rate approach, B: Bonferroni corrected P-values, δ: allelic effect and its standard error (SE), A1: minority allele, MAF: frequency of the minority allele
Table 2.
List of the genome-wide significant cis-eQTLs detected in the liver tissue (those that have been consistently detected in the muscle are shown in bold)1
| liver cis-eQTL | Gene | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSC | N | SNP | Region (Mb) | P-value | q-value | B | δ ± SE | A1 | MAF | Acronym | Region (Mb) |
| 1 | 15 | H3GA0004318 | 251.1–254.7 | 8.56E-10 | 2.43E-05 | 2.43E-05 | 0.49 ± 0.07 | A | 0.281 | HSDL2 | 253.1–253.1 |
| 1 | 1 | ASGA0006805 | 253.4–253.4 | 9.61E-07 | 2.73E-02 | 2.73E-02 | 0.22 ± 0.04 | C | 0.411 | CDC26 | 253.8–253.9 |
| 2 | 7 | MARC0045154 | 4.6–8.8 | 2.63E-06 | 3.22E-02 | 7.47E-02 | 0.27 ± 0.06 | A | 0.146 | CTSF | 5.8–5.8 |
| 2 | 17 | DIAS0003663 | 82.9–89.7 | 8.30E-09 | 1.18E-04 | 2.36E-04 | 0.30 ± 0.05 | A | 0.411 | SERINC5 | 88.8–88.9 |
| 4 | 8 | DIAS0000176 | 96.7–99.8 | 2.13E-07 | 2.27E-03 | 6.04E-03 | 0.30 ± 0.05 | A | 0.255 | APH1A | 98.8–98.8 |
| 5 | 1 | H3GA0016032 | 19.8–19.8 | 8.25E-07 | 1.17E-02 | 2.35E-02 | −0.38 ± 0.07 | G | 0.24 | PPP1R1A | 19.7–19.7 |
| 6 | 23 | ALGA0116876 | 17.0–21.7 | 9.05E-16 | 0.00E+ 00 | 0.00E+ 00 | 0.86 ± 0.08 | A | 0.255 | ARL2BP | 19.2–19.2 |
| 6 | 13 | MARC0031169 | 70.1–71.6 | 9.29E-07 | 3.30E-03 | 2.64E-02 | −0.34 ± 0.04 | G | 0.276 | LZIC | 70.2–70.2 |
| 6 | 1 | ASGA0104284 | 80.6–80.6 | 1.70E-06 | 4.82E-02 | 4.82E-02 | −0.20 ± 0.04 | C | 0.141 | ZNF436 | 81.2–81.2 |
| 6 | 14 | ALGA0114962 | 82.1–89.0 | 6.79E-06 | 1.76E-02 | 1.93E-01 | −0.32 ± 0.06 | A | 0.24 | BSDC1 | 88.8–88.8 |
| 6 | 2 | ASGA0030033 | 158.1–158.6 | 9.06E-07 | 2.57E-02 | 2.57E-02 | −0.24 ± 0.04 | G | 0.328 | YIPF1 | 158.4–158.4 |
| 7 | 2 | DRGA0007123 | 9.9–10.4 | 5.49E-10 | 1.56E-05 | 1.56E-05 | −0.40 ± 0.05 | G | 0.24 | NOL7 | 9.9–9.9 |
| 8 | 13 | ALGA0112294 | 10.6–11.3 | 3.37E-08 | 1.37E-04 | 9.58E-04 | −0.36 ± 0.06 | C | 0.406 | TAPT1 | 11.3–11.4 |
| 9 | 2 | ASGA0094801 | 46.6–46.7 | 3.33E-06 | 4.73E-02 | 9.46E-02 | −0.54 ± 0.10 | G | 0.266 | THY1 | 46.6–46.6 |
| 10 | 1 | ASGA0046443 | 10.2–10.2 | 4.07E-06 | 1.27E-02 | 1.16E-01 | −0.25 ± 0.05 | G | 0.229 | MARC2 | 10.1–10.1 |
| 10 | 10 | ALGA0106385 | 48.1–54.6 | 1.22E-07 | 2.04E-03 | 3.46E-03 | 0.24 ± 0.04 | G | 0.396 | KIAA1217 | 50.7–51.2 |
| 12 | 2 | MARC0042683 | 24.1–24.7 | 5.74E-07 | 1.63E-02 | 1.63E-02 | −0.30 ± 0.06 | G | 0.297 | PNPO | 24.1–24.2 |
| 12 | 3 | M1GA0016795 | 42.8–43.1 | 1.61E-06 | 2.29E-02 | 4.58E-02 | −0.32 ± 0.06 | A | 0.464 | CRLF3 | 42.9–42.9 |
| 12 | 9 | ALGA0066653 | 44.0–46.4 | 6.21E-12 | 9.00E-08 | 1.80E-07 | 0.31 ± 0.04 | G | 0.5 | TMEM97 | 44.6–44.6 |
| 12 | 3 | ALGA0066917 | 51.0–59.3 | 3.85E-06 | 3.65E-02 | 1.09E-01 | 0.19 ± 0.04 | G | 0.333 | RABEP1 | 51.6–51.8 |
| 13 | 1 | ALGA0070572 | 61.1–61.1 | 1.97E-06 | 2.80E-02 | 5.60E-02 | −0.40 ± 0.07 | A | 0.474 | ITPR1 | 61.0–61.3 |
| 13 | 6 | ALGA0103755 | 141.3–148.9 | 1.73E-07 | 1.52E-03 | 4.92E-03 | 0.44 ± 0.06 | A | 0.286 | GTPBP8 | 146.9–146.9 |
| 14 | 11 | H3GA0038597 | 6.5–7.2 | 1.86E-09 | 4.94E-05 | 5.28E-05 | −0.39 ± 0.06 | A | 0.401 | POLR3D | 6.5–6.5 |
| 14 | 22 | H3GA0039048 | 10.1–14.0 | 8.60E-11 | 4.10E-07 | 2.44E-06 | 0.35 ± 0.05 | G | 0.469 | HMBOX1 | 12.6–12.8 |
| 14 | 2 | DRGA0013801 | 42.7–43.4 | 2.70E-07 | 7.66E-03 | 7.66E-03 | −0.33 ± 0.06 | A | 0.406 | GRK3 | 43.3–43.4 |
| 14 | 2 | ASGA0065623 | 100.9–100.9 | 1.21E-06 | 1.24E-02 | 3.43E-02 | 0.37 ± 0.07 | A | 0.234 | LIPA | 101.1–101.2 |
| 16 | 5 | ALGA0088786 | 5.7–5.8 | 1.04E-07 | 9.88E-04 | 2.96E-03 | 0.61 ± 0.10 | G | 0.406 | RETREG1 | 5.7–5.8 |
| 18 | 8 | ASGA0100292 | 0.2–2.3 | 4.33E-06 | 3.08E-02 | 1.23E-01 | −0.15 ± 0.03 | A | 0.234 | ESYT2 | 0.5–0.6 |
1SSC: porcine chromosome, N: Number of SNPs significantly associated with the trait under study, SNP: SNP displaying the most significant association with the trait under study, Region (Mb): region containing SNPs significantly associated with the trait under study, P-value: nominal P-value, q-value: q-value calculated with a false discovery rate approach, B: Bonferroni corrected P-values, δ: allelic effect and its standard error (SE), A1: minority allele, MAF: frequency of the minority allele
Fig. 2.
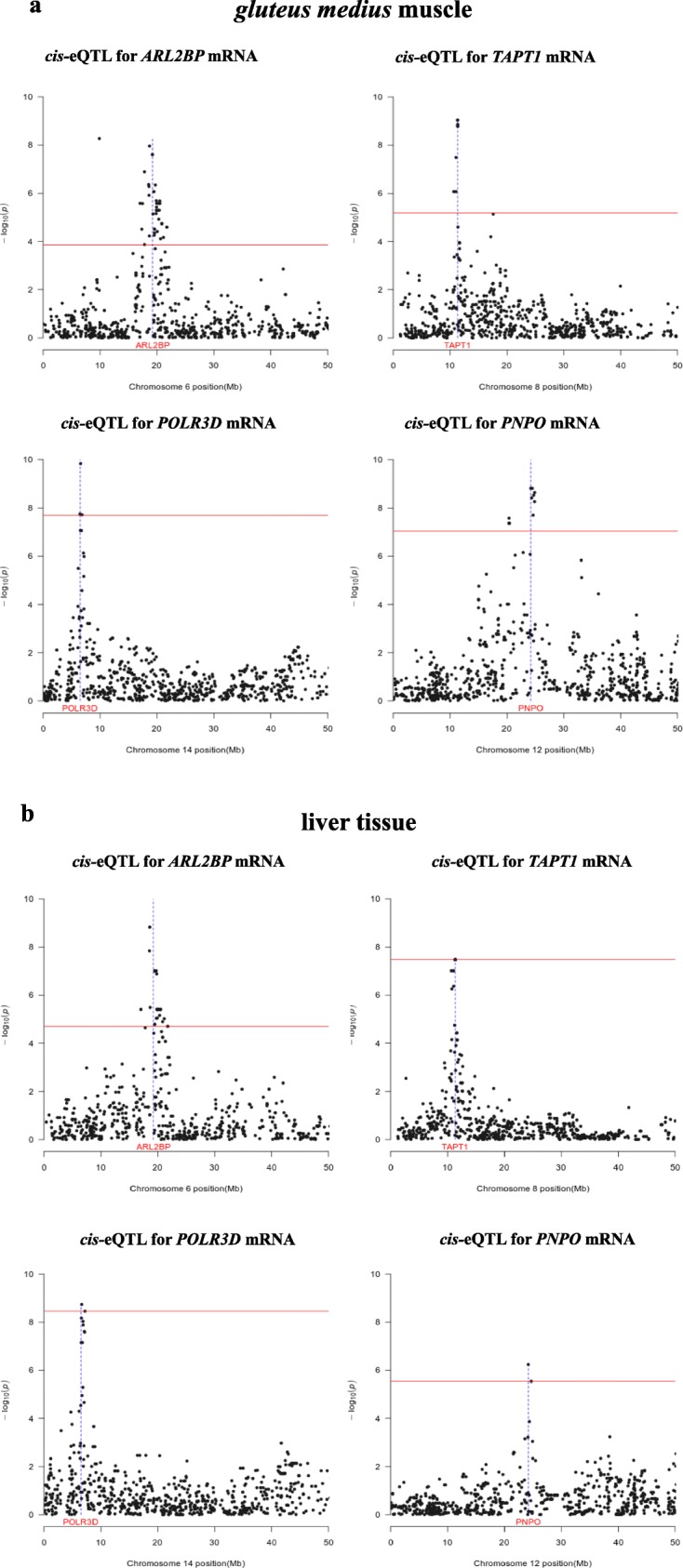
Examples of genes that are regulated by the same cis-eQTL in the gluteus medius muscle (a) and liver (b). In the Manhattan plots, the horizontal line indicates the threshold of significance after correction for multiple testing, whilst the vertical line depicts the genomic location of the four genes (ARL2BP, TAPT1, POLR3D and PNPO) under consideration
Fig. 3.
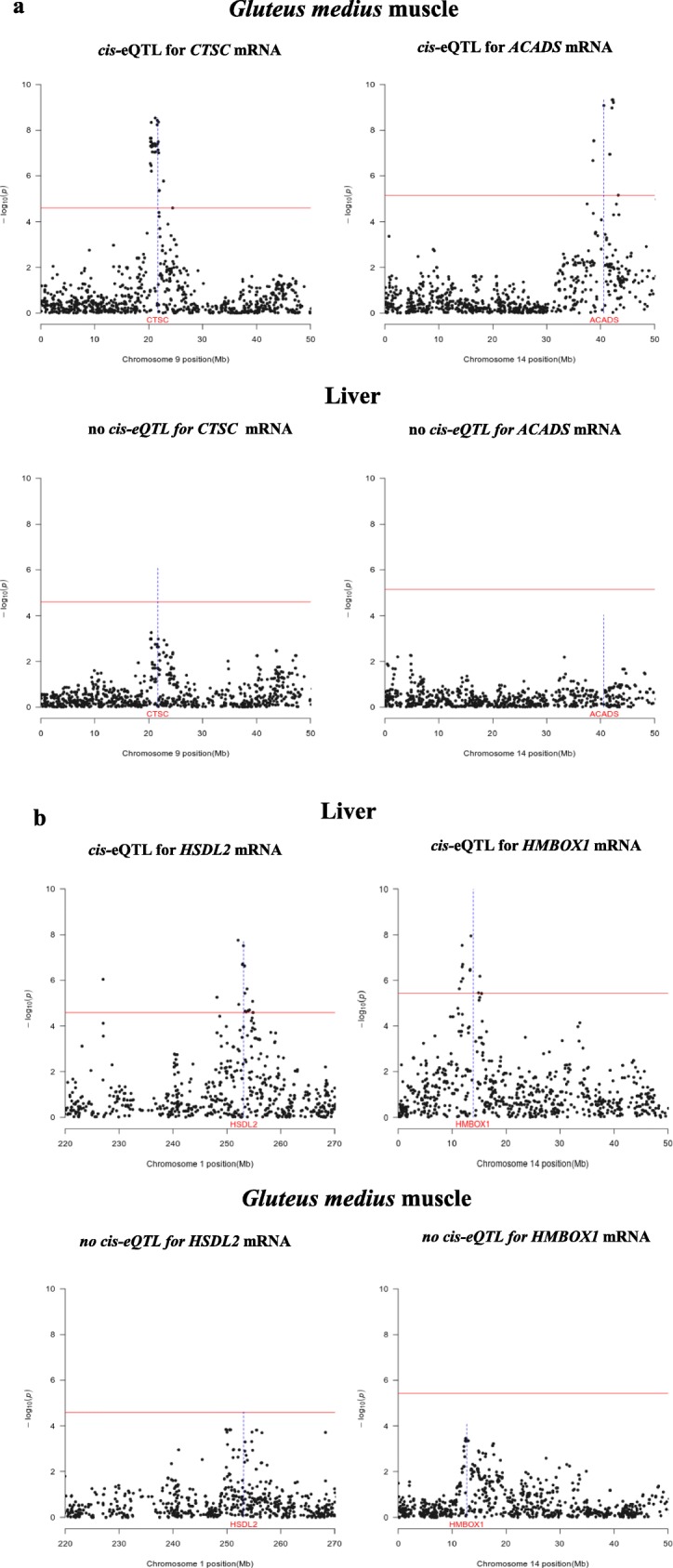
Examples of cis-eQTLs that are found in the muscle but not in the liver (a) and vice versa (b). In the Manhattan plots, the horizontal line indicates the threshold of significance after correction for multiple testing, whilst the vertical line depicts the genomic location of the CTSC, ACADS, HSDL2 and HMBOX1 genes
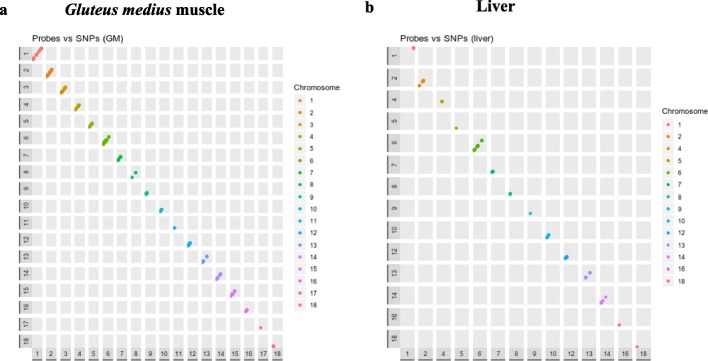
Fig. 4.
Genomic position of cis-regulated probes vs genomic positions of SNPs significantly associated with the expression of such probes in the muscle (a) and in the liver (b). Diagonal dots represents cis-eQTLs (dots from different chromosomes have different colors)
Co-localization of copy number variants and eQTLs
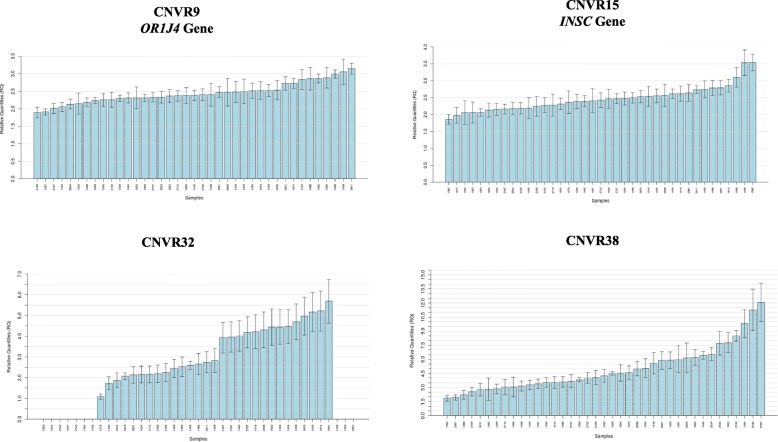
The analysis of structural variation was performed with two softwares: PennCNV and GADA. We detected 93 and 103 CNVRs with PennCNV and GADA, respectively, and 44.08% of the CNVRs detected with PennCNV were also called by GADA. These 41 common CNVRs were distributed along 13 pig chromosomes (Additional file 3). The proportions of copy gain, loss and loss/gain CNVRs were ~ 48.78%, ~ 39.02% and ~ 12.19% respectively. The size of the CNVRs ranged between 31.4 kb and 5.2 Mb, with a mean of 457.4 kb. We compared our CNVR dataset with other CNVRs previously reported in pigs [22–29], and found that 60.97% of our CNVRs had been previously reported (Additional file 4). Real time quantitative assays were designed and used to validate 4 CNVRs (CNVR 9, 15, 32 and 38) in 39 porcine samples. According to D’haene [30], estimates of copy number between 1.414 and 2.449 most likely correspond to a normal copy number of 2, whilst anything below or above these thresholds might represent a deletion (CN = 1) or a duplication (CN = 3), respectively. Following these criteria, the four regions under analysis showed evidence of structural variation (Fig. 5). The co-localization of CNVRs and eQTLs was also analyzed (Additional file 5). In the GM muscle, 2 CNVRs co-localized with 3 cis-eQTLs, while 2 CNVRs co-localized with 2 hepatic cis-eQTLs. This low concordance between CNVRs and cis-eQTLs was not surprising as only 10 CNVRs co-localized with the genomic coordinates of the gene data set analyzed in our experiment.
Fig. 5.
Relative expression values of four copy number variation regions validated by quantitative real-time PCR analysis. Each analysed individual is represented in the x-axis, while the y-axis shows the corresponding relative quantification (RQ) value. We have assigned a value of 2 to the arithmetic mean of the samples used as calibrators
Discussion
The profiles of mRNA expression of the porcine liver and skeletal muscle are highly divergent
Skeletal muscle and liver have been selected as target tissues to perform a comparative eQTL analysis because they have a key role in body energy homeostasis, a parameter that has a strong impact on growth and fat deposition in pigs. The analysis of differential expression revealed that the patterns of expression of these two tissues are highly differentiated in pigs (Fig. 1), probably as a consequence of their distinct embryonic origins and physiological functions [13]. Skeletal muscle and liver also show different patterns of gene expression in mouse [31] and humans [4]. The strongly up-regulated genes in muscle included BIN1, which mediates calcium signaling and excitation–contraction coupling [32], tropomyosin (TPM2), which is fundamental for muscle contraction [33], and ZNF106, which is required for the postnatal maintenance of myofiber innervation by motor neurons [34]. In the liver, we detected a strong overexpression of SORD, an enzyme that converts sorbitol into fructose [35], HSD17B13, whose inactivation leads to chronic liver disease [36], and APOA1, the major structural component of high-density lipoproteins [37].
Identification of cis-eQTLs with consistent effects on the mRNA levels of genes expressed in the liver and skeletal muscle
In pigs, hundreds of eQTLs with effects on the muscle transcriptome have been detected, but the majority of these studies focused on target subsets of genes either mapping to QTLs [12] or displaying significant expression-phenotype correlations [5, 6, 10, 11]. Herewith, we have detected 76 muscle cis-eQTLs and 28 liver cis-eQTLs which attained genome-wide significance. These numbers are consistent with previous reports. Liaubet et al. [38] made a genome scan based on microarray measurements of longissimus lumborum gene expression in 57 pigs and identified 335 eQTLs. Of these, only 18 had cis-regulatory effects. Similarly, Cánovas et al. [8] identified 478 skeletal muscle genome-wide significant eQTLs but only 13% acted in cis-. Although modest sample size (N = 103) limits our ability to identify eQTLs, this sample size ought to allow detecting eQTLs with moderate to large effects on gene expression. As a valid reference, the Genotype-Tissue Expression project detected thousands of eQTLs in 44 tissues characterized by RNA-Seq, and 18 of these tissues were represented by sample sizes equal or below the one used in the current experiment [2]. However, it is also true that sample size largely affects the statistical power of eQTL mapping [4] and thus, the analysis of a larger number of pigs would most likely uncover additional eQTLs that remained undetected in our study.
Our results indicate that almost 42.8% of the cis-eQTLs regulating gene expression in the liver also display significant associations with the muscle mRNA levels of the very same genes (Tables 1 and 2, Fig. 2). The liver and muscle common cis-eQTLs encompass genes previously related to the function of these two organs, such as the cathepsin F (CTSF) gene, that harbors a polymorphism that has been associated with longissimus dorsi tenderness, ham weight and fatness in Italian crossbred pigs [39], the transmembrane anterior posterior transformation 1 (TAPT1) gene, which was linked to carcass and eviscerated weight in a GWAS in chicken [40], the mitochondrial amidoxime reducing component 2 (MARC2) gene, that appears to be involved in porcine fatness [41], and the transmembrane protein 97 (TMEM97) gene, which contributes to the regulation of cholesterol levels [42].
There is also a relevant fraction of porcine cis-eQTLs that display significant effects only in one of the two tissues (Figs. 3 and 4). In the Genotype-Tissue Expression (GTEx) pilot experiment [4], approximately 50% of eQTLs were shared by the nine human tissues under analysis. Moreover, two main types of eQTLs were especially prevalent, i.e. those that regulate gene expression in a single tissue and those that are ubiquitously detected in all tissues. Interestingly, the GTEx pilot analysis also showed that eQTLs affecting gene expression in the skeletal muscle show a limited replicability in other tissues [4]. In other studies, also performed in humans, the eQTL tissue-specificity ranged between 50% [43] and 60–80% [44], which implies that the effects of many regulatory mutations are modulated by tissue-associated factors.
Several of the cis-eQTLs detected in our experiment affected the expression of genes involved in lipid or carbohydrate metabolism. For instance, the down-regulation of HTATIP2 leads to elevated fatty acid synthesis and enhanced levels of lipogenic enzymes [14]. The EXOC7 gene regulates the uptake of fatty acids by adipocytes and ACADS is involved in the ß-oxidation of fatty acids [16], while FAM3C can suppress hepatic gluconeogenesis [45]. It would be interesting to investigate whether polymorphisms associated with the expression of lipid genes also display associations with fatness traits. Two of the muscle cis-eQTLs detected in our study have been previously reported by other authors. An eQTL for the BSDC1 gene was detected by Ponsuksili et al. [46] and the expression of this gene was also correlated with the percentage of weight loss of the longissimus dorsi muscle. Moreover, a local eQTL that regulates the expression of PNPO and which co-localizes with several meat quality retail traits (such as the percentage of fat and moisture in meat) was described by Steibel et al. [7]. A remarkable level of heterogeneity has been observed in the genetic determinism of production traits in different porcine breeds [47]. In consequence, we anticipated a limited positional concordance amongst eQTLs detected in different breeds. Indeed, a joint analysis of eQTLs across five human populations revealed that varying linkage disequilibrium patterns across populations results in the detection of large numbers of eQTLs with heterogeneous effects [48].
Limited positional concordance between cis-eQTLs and copy number variation regions
Another goal of our study was to investigate the co-localization of cis-eQTLs and CNVRs in order to make an initial assessment of the potential impact of structural variation on gene expression in pigs. With PennCNV and GADA, we consistently detected 41 CNVRs with a mean size of 457.4 kb. When we compared our CNVR dataset with CNVRs previously reported in pigs [22–29], we found that 60.97% of the CNVRs detected by us had been previously reported in the literature (Additional file 4). A moderate agreement of CNVR locations amongst studies and populations has been evidenced in several reports [27, 49–52]. Discrepancies could be due to differences in the genetic background of the populations under analysis, filtering criteria (correction factors, criteria to define CNV and CNVRs, etc.), genotyping methods, CNV calling algorithms and the use of family information [27, 53]. In the GM muscle, 2 CNVRs (14 and 38) co-localized with 2 cis-eQTLs (SSC2, 32.6–39.4 Mb and SSC14, 102.7–106.9 Mb), whilst the co-localizations between CNVRs and cis-eQTLs in the liver tissue were somewhat similar, i.e. 2 cis-eQTLs (SSC2, 5.83–5.90 Mb and SSC13, 148.90–149.17 Mb) co-localized with CNVR 11 and 36. This limited positional concordance is probably due to the fact that only 10 CNVRs co-localized with the set of probes analyzed in the current experiment. The low positional concordance between CNVRs and eQTLs could be also due to technical reasons related to the difficulties of detecting CNVs with SNP arrays and the limited sensitivity and poor annotation of porcine microarrays. Besides, the search of CNVs was performed in a population of 350 individuals, while only 103 pigs had available gene-expression data. On the other hand, variations in copy number do not necessarily involve changes in gene expression, e.g. in heterozygous individuals, the loss of one allele can be compensated by an increase in the expression of the other allele, and duplication can generate additional copies which expression is silenced [53]. A future objective would be to investigate the association between the number of copies of genes and their expression levels, but a high throughput method allowing the accurate determination of CNV genotypes will be needed to achieve this goal (genotype determination based on 60 K SNP array data is too imprecise to carry out such analyses).
Conclusions
Despite the fact that there are considerable differences in the gene expression patterns of the porcine liver and skeletal muscle, we have identified a substantial proportion of common cis-eQTLs regulating gene expression in both tissues.
Materials and methods
Phenotyping and genotyping of a commercial Duroc population
We have used a commercial Duroc line of 350 Duroc barrows that were slaughtered at the age of ~ 190 days, with an approximate live weight of 122 kg. This population was generated by crossing 5 boars with ~ 400 sows and selecting one offspring/sow (50 piglets did not complete their productive cycle so we ended up with 350 barrows with valid records). The 350 barrows were bred in the experimental farm of the Pig Control Center of the Institut de Recerca i Tecnologia Agroalimentàries (IRTA). The specific conditions of management and feeding have been previously reported [54, 55]. Pigs were slaughtered in a commercial abattoir following the guidelines of the Spanish Royal Decree 54/1995 (January 20, 1995) to preserve animal welfare. In this way, swine were stunned with carbon dioxide (70% or higher) until they lost consciousness and they were subsequently bled by making an incision in, at least, one carotid artery. At slaughter, GM muscle and liver biopsies were obtained for 103 pigs. Total RNA purification procedures have been previously reported [8, 56]. All experimental procedures were approved by the Ethical Committee of IRTA.
Genomic DNA was extracted from blood samples by following a standard phenol-chloroform protocol. Each pig was genotyped for 62,163 SNPs with the Porcine SNP60 BeadChip (Illumina, SanDiego, CA). The quality of the genotyping results was evaluated with the GenomeStudio software (Illumina). The PLINK software [57] was used to filter out the SNP markers with minor allele frequency below 5%, missing genotype rate above 10%, and displaying significant departures (P-value < 0.001) from the Hardy-Weinberg equilibrium. Markers which could not be mapped to the Sscrofa11.1 assembly or that mapped to either the X or Y chromosomes were also eliminated from the data set. Markers in complete linkage disequilibrium (r2 > 0.98) were also purged. The final data set used for performing eQTL analyses contained 28,493 SNPs. The filtering criteria used for the CNV analysis was different and only SNPs that did not map to the Sscrofa11.1 assembly or that were located in sex chromosomes were removed, leading to a final number of 46,537 used SNPs.
Differential expression analysis between muscle and liver tissue
Gluteus medius muscle and liver samples were collected from 103 Duroc pigs (Lipgen population) after slaughtering, and immediately frozen in liquid nitrogen. These 103 pigs were selected on the basis of a principal component analysis focused on 13 lipid and growth related traits [58]. We chose individuals representing two different metabolic types, i.e. (i) fat pigs with high intramuscular fat (high saturated and monounsaturated fatty acid content) and also high serum lipid levels, and (ii) pigs that were lean and displayed a low level of intramuscular fat (high polyunsaturated fatty acid content) and circulating lipids [58]. Total RNA was extracted from both GM and liver samples, and mRNA expression profiles were characterized by hybridization to the GeneChip Porcine arrays (Affymetrix Inc., Santa Clara, CA), as previously reported by Cánovas et al. [58]. Hepatic and muscular microarray expression data were deposited in the Gene Expression Omnibus (GEO) public repository, and are accessible through GEO Series accession number GSE115484. The Robust Multi-array Average (RMA) algorithm [59] was employed for carrying out data pre-processing, background correction, normalization and log-transformation of expression values. Gene Intensity level of significance for detecting expressed probes was calculated with the MAS 5.0 algorithm [60]. Control probes and those probes that did not show expression levels above the detection threshold in all samples were filtered out. Differential expression analysis between the GM muscle and liver followed the guidelines of the limma-trend pipeline [61, 62]. The limma’s empirical Bayes method incorporated a mean-variance trend, thus making possible to adequately model the relationship between variance and gene signal intensity. In our study, fold-change (FC) values reflect the mean expression levels in the GM muscle vs liver. The correspondence between differentially expressed probes (|FC| > 1.5; q-value < 0.05) and genes was based on the Affymetrix porcine annotation data assembled database [63] and the Biomart database (https://www.ensembl.org/biomart/martview).
Genome scan for expression QTLs
The genome scan for eQTLs targeted a total of 3326 probes that are simultaneously expressed in both tissues. These genes were selected using the following criteria: The MAS 5.0 [60] algorithm was used to detect the probes with expression levels above the Gene Intensity level of significance. Probes were retained when expressed in all the samples analyzed in both tissues. Expressed probes where then annotated according to Biomart database available at Ensembl repositories (https://www.ensembl.org/biomart/martview) and those with no gene correspondence were removed from further analyses. We used the GEMMA software [64] and followed the methods described by González-Prendes et al. [56] to carry out the association analyses. The fixed effects and parameters assumed in the statistical model were:
where y is the vector that defines the expression of each gene in the GM muscle and liver of the ith individual; W is the matrix with a column of 1 s and the fixed effects, i.e.“batch of fattening” (with 4 categories) and “laboratory” (microarray data were generated in two different laboratories); α is a c-vector of the corresponding coefficients including the intercept; x is an n-vector of marker genotypes; β is the SNP allelic effect estimated as a regression coefficient on the corresponding x genotype (values − 1, 0, 1); u is a n-vector of random effects with a n-dimensional multivariate normal distribution MVNn (0, λ τ − 1 K) where τ− 1 is the variance of the residual errors; λ is the ratio between the two variance components; K is a known relatedness matrix derived from SNPs and ε is the vector of errors with an MVNn (0, τ − 1 I n) being In the identity matrix. The false discovery rate approach reported by Benjamini and Hochberg [65] was used to correct for multiple testing at the genome-wide level. In this study, cis-eQTLs were defined as these genomic regions with SNPs significantly associated with a probe located at a maximum distance of ±1 Mb of the associated SNPs. Co-localization was defined as an overlap of at least 1 base pair between the genomic locations of two eQTLs. The GM muscle cis-eQTLs detected with microarrays were validated by considering an RNA-seq data set corresponding to the same tissue and comprising 52 Duroc pigs [66]. These 52 pigs were a subset of the 103 pigs investigated in the current work and the methods to generate GM muscle RNA-seq data are fully described in [66].
Detection of copy number variation
The PennCNV software [67] was employed to detect copy number variants (CNVs) on the basis of the information provided by 46,537 autosomal SNPs. The PennCNV software implements a hidden Markov model (HMM) to infer CNV calls for each genotyped sample using as input the intensity signal Log R Ratio (LRR) and the B Allele Frequency (BAF) information generated with the BeadStudio software (Illumina). Samples with a standard deviation of LRR > 0.30 and BAF drift > 0.01 were discarded. Besides, a wave adjustment procedure for genomic waves was carried out [67]. With these filtering steps, 20 samples were eliminated from the data set. Only CNVs spanning three or more consecutive SNPs were taken into account. The CNV calling was performed using the default parameters of the HMM model by assuming a UF factor of 0.01. Copy number variant regions (CNVRs) were created by merging CNVs with an overlap of 80% or more. Additionally we used the Genome Alteration Detection Analysis (GADA) package [68] to further validate the CNVR detected with PennCNV and minimize the rate of false positives. The GADA software uses a sparse Bayesian algorithm, based on LRR and BAF values obtained along the genome with the BeadStudio software (Illumina), to identify genomic locations where copy number changes occur. Briefly, a LRR average of markers in a chromosome segment is computed and compared to the median of the respective chromosome. Finally, the copy number class is classified as gain, loss or gain/loss. In our study, copy number variations spanning three or more consecutive SNPs were taken into account and the multiple array analysis option was employed. The parameters defined for the Bayesian learning model and the backward elimination were: 0.8 for the sparseness hyperparameter and 8 for the critical value of the backward elimination.
Validation of copy number variant regions by quantitative PCR
Quantitative real time PCR (qPCR) assays were used to validate four CNVRs in 39 porcine samples randomly selected from the Lipgen population. The relative quantification (RQ) of the CNVRs was done as previously described [24]. Primers (Additional file 6) were designed with the Primer Express Software (Applied Biosystems). Copy number variant regions were quantified in 384-well plates using SYBR Select Master Mix in a QuantStudio 12 K Flex Real-Time PCR System platform (Applied Biosystems, Inc., Foster City, CA). Reactions were performed in triplicate, and they contained 7.5 ng of genomic DNA and primers at 300 nM in a final volume of 15 μl. The thermocycling profile was: one cycle at 95 °C for 10 min plus 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Moreover, a melting curve profile (95 °C for 15 s, 60 °C for 15 s and a gradual increase in temperature with a ramp rate of 1% up to 95 °C) was implemented to maximize the specificity of the amplification reactions. Relative expression values were calculated with the Qbase+ software (Biogazelle, Ghent, Belgium) by applying the 2-ΔΔCt method (after verifying that its assumptions were adequately fulfilled) [69]. Relative expression values were calibrated using the arithmetic mean of 3–5 samples showing the lowest number of copies for each specific assay. In the specific case of CNVR32, which encompasses a deletion, the five samples chosen for calibration were those with RQ values around 2. Normalization of the expression data was done by using a previously reported assay based on the glucagon gene [70].
Additional files
Table S1. List of genes that are differentially expressed (DE) in the gluteus medius muscle and the liver (q-value < 0.05). (XLSX 353 kb)
Figure S1. Boxplots depicting the mRNA expression levels of cis-eQTL regulated genes measured with RNA-Seq and microarrays in the gluteus medius muscle of 52 and 103 Duroc pigs, respectively. Means were compared with a Student’s t- test: P-value > 0.05 (ns); P-value ≤ 0.05 (*); P-value ≤ 0.01 (**); P-value ≤ 0.001 (***) and P-value ≤ 0.0001 (****). (DOCX 807 kb)
Table S2. Locations, absolute frequencies, lengths and status of CNVRs detected in 350 Duroc pigs with the PennCNV and GADA softwares. (XLSX 12 kb)
Table S3. Positional coincidence of the copy number variant regions (CNVR) detected in the current work with CNVRs reported in other pig populations. (XLSX 10 kb)
Table S4. Co-localization of CNVRs and cis-eQTLs in the gluteus medius muscle (eQTL and gene positions are expressed in Mb). Table S5. Co-localization of CNVRs and cis-eQTLs in liver tissue (eQTL and gene positions are expressed in Mb). (XLSX 13 kb)
Table S6. Sequences of the primers used to validate CNVRs by qPCR. (XLSX 9 kb)
Acknowledgements
The authors are indebted to Selección Batallé S.A. for providing the animal material. We gratefully acknowledge to J. Reixach (Selección Batallé) and J. Soler (IRTA) for their collaboration in the experimental protocols.
Abbreviations
- |FC|
absolute value of the fold-change
- bp
base pair
- CNV
Copy number variant
- CNVR
Copy number variant regions
- DE
Differentially expressed
- e.g.
for example
- eQTL
expression quantitative trait loci
- GEMMA
Genome-wide Efficient Mixed-Model Association
- GEO
Gene Expression Omnibus
- GM
gluteus medius
- GWAS
genome-wide association analysis
- i.e.
in other words
- Kb
Kilobase pairs
- Mb
Megabase pairs
- mRNA
messenger RNA
- SNP
Single nucleotide polymorphism
- SSC
porcine chromosome
Authors’ contributions
MA and RQ designed the experiment; AC1 and AM contributed to generate SNP data; EMS made the analyses of differential expression and selected the genes tested in the eQTL analysis; RGP and AC2 analyzed the eQTL data; RGP analyzed the CNV data; AZ and YRC provided support in the analysis of CNV data; AC1 and RGP validated CNV data by quantitative PCR; TFC and AC2 provided bioinformatic support in the analysis of gene expression data; MA and RGP wrote the paper; all authors read and approved the manuscript.
Funding
Part of the research presented in this publication was funded by grants AGL2013–48742-C2–1-R and AGL2013–48742-C2–2-R awarded by the Spanish Ministry of Economy and Competitivity. We also acknowledge the support of the Spanish Ministry of Economy and Competitivity for the Center of Excellence Severo Ochoa 2016–2019 (SEV-2015-0533) grant awarded to the Centre for Research in Agricultural Genomics. Ángela Cánovas was funded under the Juan de la Cierva program awarded by the Spanish Ministry of Science. Taina F Cardoso was funded with a fellowship from the CAPES Foundation-Coordination of Improvement of Higher Education, Ministry of Education (MEC) of the Federal Government of Brazil. Rayner González-Prendes was funded by a FPU Ph.D. grant from the Spanish Ministry of Education (FPU12/00860). Emilio Mármol-Sánchez was funded by a FPU Ph.D. grant from the Spanish Ministry of Education (FPU15/01733).
Availability of data and materials
Microarray expression data can be found in 10.6084/m9.figshare.7952054, while Porcine SNP60 BeadChip data can be accessed at 10.6084/m9.figshare.7473092.v2
Ethics approval
Animal care, management procedures and blood sampling were performed following national guidelines for the Good Experimental Practices and they were approved by the Ethical Committee of the Institut de Recerca i Tecnologia Agroalimentàries (IRTA).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rayner González-Prendes and Emilio Mármol-Sánchez share the first authorship.
References
- 1.Nica A. C., Dermitzakis E. T. Expression quantitative trait loci: present and future. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1620):20120362–20120362. doi: 10.1098/rstb.2012.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 3.Gamazon ER, Nicolae DL, Cox NJ. A study of CNVs as trait-associated polymorphisms and as expression quantitative trait loci. PLoS Genet. 2011;7:e1001292. doi: 10.1371/journal.pgen.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardlie KG, Deluca DS, Segre AV, Sullivan TJ, Young TR, Gelfand ET, et al. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, et al. Trait correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics. 2008;9:367. doi: 10.1186/1471-2164-9-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wimmers K, Murani E, Ponsuksili S. Functional genomics and genetical genomics approaches towards elucidating networks of genes affecting meat performance in pigs. Brief Funct Genomics. 2010;9:251–258. doi: 10.1093/bfgp/elq003. [DOI] [PubMed] [Google Scholar]
- 7.Steibel JP, Bates RO, Rosa GJM, Tempelman RJ, Rilington VD, Ragavendran A, et al. Genome-wide linkage analysis of global gene expression in loin muscle tissue identifies candidate genes in pigs. PLoS One. 2011;6:e16766. doi: 10.1371/journal.pone.0016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cánovas A, Pena RN, Gallardo D, Ramírez O, Amills M, Quintanilla R. Segregation of regulatory polymorphisms with effects on the gluteus medius transcriptome in a purebred pig population. PLoS One. 2012;7:e35583. doi: 10.1371/journal.pone.0035583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidt H, Cinar MU, Uddin MJ, Looft C, Jüngst H, Tesfaye D, et al. A genetical genomics approach reveals new candidates and confirms known candidate genes for drip loss in a porcine resource population. Mamm Genome. 2013;24:416–426. doi: 10.1007/s00335-013-9473-z. [DOI] [PubMed] [Google Scholar]
- 10.Ponsuksili S, Murani E, Brand B, Schwerin M, Wimmers K. Integrating expression profiling and whole-genome association for dissection of fat traits in a porcine model. J Lipid Res. 2011;52:668–678. doi: 10.1194/jlr.M013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Yang B, Zeng Z, Yang H, Liu C, Ren J, et al. Genetic dissection of blood lipid traits by integrating genome-wide association study and gene expression profiling in a porcine model. BMC Genomics. 2013;14:848. doi: 10.1186/1471-2164-14-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz M, Rodríguez MC, Alves E, Folch JM, Ibañez-Escriche N, Silió L, et al. Genome-wide analysis of porcine backfat and intramuscular fat fatty acid composition using high-density genotyping and expression data. BMC Genomics. 2013;14:845. doi: 10.1186/1471-2164-14-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferraz AL, Ojeda A, López-Béjar M, Fernandes LT, Castelló A, Folch JM, et al. Transcriptome architecture across tissues in the pig. BMC Genomics. 2008;9:173. doi: 10.1186/1471-2164-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin F, Sharen G, Yuan F, Peng Y, Chen R, Zhou X, et al. TIP30 regulates lipid metabolism in hepatocellular carcinoma by regulating SREBP1 through the Akt/mTOR signaling pathway. Oncogenesis. 2017;6:e347. doi: 10.1038/oncsis.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue M, Akama T, Jiang Y, Chun TH. The exocyst complex regulates free fatty acid uptake by adipocytes. PLoS One. 2015;10:e0120289. doi: 10.1371/journal.pone.0120289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Kruger C, Wicks S, Simon J, Kumar KG, Johnson WD, et al. Short chain acyl-CoA dehydrogenase deficiency and short-term high-fat diet perturb mitochondrial energy metabolism and transcriptional control of lipid-handling in liver. Nutr Metab. 2016;13:17. doi: 10.1186/s12986-016-0075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E. A., Frosch M. P., Eckman C. B., Tanzi R. E., Selkoe D. J., Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid -protein, and the -amyloid precursor protein intracellular domain in vivo. Proceedings of the National Academy of Sciences. 2003;100(7):4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle RE, Mydock-McGrane L, et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9–Niemann-pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Ding L, Yang W, Wang J, Chen L, Chang Y, et al. Hepatic activation of the FAM3C-HSF1-CaM pathway attenuates hyperglycemia of obese diabetic mice. Diabetes. 2017;66:1185–1197. doi: 10.2337/db16-0993. [DOI] [PubMed] [Google Scholar]
- 20.Skogsberg J, Lundström J, Kovacs A, Nilsson R, Noori P, Maleki S, et al. Transcriptional profiling uncovers a network of cholesterol-responsive atherosclerosis target genes. PLoS Genet. 2008;4:e1000036. doi: 10.1371/journal.pgen.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubland JA, Francis GA. Lysosomal acid lipase: at the crossroads of normal and atherogenic cholesterol metabolism. Front Cell Dev Biol. 2015;3:3. doi: 10.3389/fcell.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fadista J, Nygaard M, Holm L-E, Thomsen B, Bendixen C. A snapshot of CNVs in the pig genome. PLoS One. 2008;3:e3916. doi: 10.1371/journal.pone.0003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler KE, Pong-Wong R, Bauer J, Clemente EJ, Reitter CP, Affara NA, et al. Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds. BMC Genomics. 2013;14:784. doi: 10.1186/1471-2164-14-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramayo-Caldas Y, Castelló A, Pena RN, Alves E, Mercadé A, Souza CA, et al. Copy number variation in the porcine genome inferred from a 60 k SNP BeadChip. BMC Genomics. 2010;11:593. doi: 10.1186/1471-2164-11-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiavo G, Dolezal MA, Scotti E, Bertolini F, Calò DG, Galimberti G, et al. Copy number variants in Italian large white pigs detected using high-density single nucleotide polymorphisms and their association with back fat thickness. Anim Genet. 2014;45:745–749. doi: 10.1111/age.12180. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Tang Z, Sun Y, Wang H, Wang C, Yu S, et al. Analysis of genome-wide copy number variations in Chinese indigenous and Western pig breeds by 60 K SNP genotyping arrays. PLoS One. 2014;9:e106780. doi: 10.1371/journal.pone.0106780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández AI, Barragán C, Fernández A, Rodríguez MC, Villanueva B. Copy number variants in a highly inbred Iberian porcine strain. Anim Genet. 2014;45:357–366. doi: 10.1111/age.12137. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Qiao R, Wei R, Guo Y, Ai H, Ma J, et al. A comprehensive survey of copy number variation in 18 diverse pig populations and identification of candidate copy number variable genes associated with complex traits. BMC Genomics. 2012;13:733. doi: 10.1186/1471-2164-13-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Mei S, Zhang X, Peng X, Liu G, Tao H, et al. Identification of genome-wide copy number variations among diverse pig breeds by array CGH. BMC Genomics. 2012;13:725. doi: 10.1186/1471-2164-13-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’haene B, Vandesompele J, Hellemans J. Accurate and objective copy number profiling using real-time quantitative PCR. Methods. 2010;50:262–270. doi: 10.1016/j.ymeth.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, et al. A comprehensive mouse transcriptomic bodymap across 17 tissues by RNA-seq. Sci Rep. 2017;7:4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Hong T. BIN1 regulates dynamic t-tubule membrane. Biochim Biophys Acta. 2016;1863:1839–1847. doi: 10.1016/j.bbamcr.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seymour J, O’Brien EJ. The position of tropomyosin in muscle thin filaments. Nature. 1980;283:680–682. doi: 10.1038/283680a0. [DOI] [PubMed] [Google Scholar]
- 34.Anderson Douglas M., Cannavino Jessica, Li Hui, Anderson Kelly M., Nelson Benjamin R., McAnally John, Bezprozvannaya Svetlana, Liu Yun, Lin Weichun, Liu Ning, Bassel-Duby Rhonda, Olson Eric N. Severe muscle wasting and denervation in mice lacking the RNA-binding protein ZFP106. Proceedings of the National Academy of Sciences. 2016;113(31):E4494–E4503. doi: 10.1073/pnas.1608423113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Kabbani O, Darmanin C, Chung RPT. Sorbitol dehydrogenase: structure, function and ligand design. Curr Med Chem. 2004;11:465–476. doi: 10.2174/0929867043455927. [DOI] [PubMed] [Google Scholar]
- 36.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamanian-Daryoush M, Lindner D, Tallant TC, Wang Z, Buffa J, Klipfell E, et al. The cardioprotective protein apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol Chem. 2013;288:21237–21252. doi: 10.1074/jbc.M113.468967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liaubet L, Lobjois V, Faraut T, Tircazes A, Benne F, Iannuccelli N, et al. Genetic variability of transcript abundance in pig peri-mortem skeletal muscle: eQTL localized genes involved in stress response, cell death, muscle disorders and metabolism. BMC Genomics. 2011;12:548. doi: 10.1186/1471-2164-12-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davoli R, Schivazappa C, Zambonelli P, Braglia S, Rossi A, Virgili R. Association study between single nucleotide polymorphisms in porcine genes and pork quality traits for fresh consumption and processing into Italian dry-cured ham. Meat Sci. 2017;126:73–81. doi: 10.1016/j.meatsci.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 40.Liu R, Sun Y, Zhao G, Wang F, Wu D, Zheng M, et al. Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in Beijing-you chickens. PLoS One. 2013;8:e61172. doi: 10.1371/journal.pone.0061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogelman LJA, Cirera S, Zhernakova DV, Fredholm M, Franke L, Kadarmideen HN. Identification of co-expression gene networks, regulatory genes and pathways for obesity based on adipose tissue RNA sequencing in a porcine model. BMC Med Genet. 2014;7:57. doi: 10.1186/1755-8794-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 44.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Wang J, Yang W, Chen J, Meng Y, Feng B, et al. FAM3C activates HSF1 to suppress hepatic gluconeogenesis and attenuate hyperglycemia of type 1 diabetic mice. Oncotarget. 2017;8:106038–106049. doi: 10.18632/oncotarget.22524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponsuksili S, Murani E, Schwerin M, Schellander K, Wimmers K. Identification of expression QTL (eQTL) of genes expressed in porcine M. longissimus dorsi and associated with meat quality traits. BMC Genomics. 2010;11:572. doi: 10.1186/1471-2164-11-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, Yang B, Zhang J, Cui L, Ma J, Chen C, et al. Genome-wide association studies for fatty acid metabolic traits in five divergent pig populations. Sci Rep. 2016;6:24718. doi: 10.1038/srep24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen X, Luca F, Pique-Regi R. Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet. 2015;11:e1005176. doi: 10.1371/journal.pgen.1005176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paudel Y, Madsen O, Megens H-J, Frantz LAF, Bosse M, Bastiaansen JWM, et al. Evolutionary dynamics of copy number variation in pig genomes in the context of adaptation and domestication. BMC Genomics. 2013;14:449. doi: 10.1186/1471-2164-14-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Jian, Li Rongrong, Li Sheng, Ran Xueqin, Wang Jiafu, Jiang Jicai, Zhao Pengju. Identification of Copy Number Variations in Xiang and Kele Pigs. PLOS ONE. 2016;11(2):e0148565. doi: 10.1371/journal.pone.0148565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiedmann Ralph T., Nonneman Dan J., Rohrer Gary A. Genome-Wide Copy Number Variations Using SNP Genotyping in a Mixed Breed Swine Population. PLOS ONE. 2015;10(7):e0133529. doi: 10.1371/journal.pone.0133529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paudel Y, Madsen O, Megens H-J, Frantz LAF, Bosse M, Crooijmans RPMA, et al. Copy number variation in the speciation of pigs: a possible prominent role for olfactory receptors. BMC Genomics. 2015;16:330. doi: 10.1186/s12864-015-1449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clop A, Vidal O, Amills M. Copy number variation in the genomes of domestic animals. Anim Genet. 2012;43:503–517. doi: 10.1111/j.1365-2052.2012.02317.x. [DOI] [PubMed] [Google Scholar]
- 54.Gallardo D, Pena RN, Amills M, Varona L, Ramírez O, Reixach J, et al. Mapping of quantitative trait loci for cholesterol, LDL, HDL, and triglyceride serum concentrations in pigs. Physiol Genomics. 2008;35:199–209. doi: 10.1152/physiolgenomics.90249.2008. [DOI] [PubMed] [Google Scholar]
- 55.Gallardo D, Quintanilla R, Varona L, Díaz I, Ramírez O, Pena RN, et al. Polymorphism of the pig acetyl-coenzyme a carboxylase α gene is associated with fatty acid composition in a Duroc commercial line. Anim Genet. 2009;40:410–417. doi: 10.1111/j.1365-2052.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- 56.González-Prendes R, Quintanilla R, Cánovas A, Manunza A, Figueiredo-Cardoso T, et al. Joint QTL mapping and gene expression analysis identify positional candidate genes influencing pork quality traits. Sci Rep. 2017;7:39830. [DOI] [PMC free article] [PubMed]
- 57.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cánovas A, Quintanilla R, Amills M, Pena RN. Muscle transcriptomic profiles in pigs with divergent phenotypes for fatness traits. BMC Genomics. 2010;11:372. doi: 10.1186/1471-2164-11-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 60.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 61.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carlson M. Affymetrix porcine annotation data (chip porcine). R package version 3.2.3. 2016. [Google Scholar]
- 64.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 66.Cardoso TF, Cánovas A, Canela-Xandri O, González-Prendes R, Amills M, Quintanilla R. RNA-seq based detection of differentially expressed genes in the skeletal muscle of Duroc pigs with distinct lipid profiles. Sci Rep. 2017;7:40005. doi: 10.1038/srep40005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pique-Regi R, Monso-Varona J, Ortega A, Seeger RC, Triche TJ, Asgharzadeh S. Sparse representation and Bayesian detection of genome copy number alterations from microarray data. Bioinformatics. 2008;24:309–318. doi: 10.1093/bioinformatics/btm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 70.Revilla M, Puig-Oliveras A, Castelló A, Crespo-Piazuelo D, Paludo E, Fernández AI, et al. A global analysis of CNVs in swine using whole genome sequence data and association analysis with fatty acid composition and growth traits. PLoS One. 2017;12:e0177014. doi: 10.1371/journal.pone.0177014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of genes that are differentially expressed (DE) in the gluteus medius muscle and the liver (q-value < 0.05). (XLSX 353 kb)
Figure S1. Boxplots depicting the mRNA expression levels of cis-eQTL regulated genes measured with RNA-Seq and microarrays in the gluteus medius muscle of 52 and 103 Duroc pigs, respectively. Means were compared with a Student’s t- test: P-value > 0.05 (ns); P-value ≤ 0.05 (*); P-value ≤ 0.01 (**); P-value ≤ 0.001 (***) and P-value ≤ 0.0001 (****). (DOCX 807 kb)
Table S2. Locations, absolute frequencies, lengths and status of CNVRs detected in 350 Duroc pigs with the PennCNV and GADA softwares. (XLSX 12 kb)
Table S3. Positional coincidence of the copy number variant regions (CNVR) detected in the current work with CNVRs reported in other pig populations. (XLSX 10 kb)
Table S4. Co-localization of CNVRs and cis-eQTLs in the gluteus medius muscle (eQTL and gene positions are expressed in Mb). Table S5. Co-localization of CNVRs and cis-eQTLs in liver tissue (eQTL and gene positions are expressed in Mb). (XLSX 13 kb)
Table S6. Sequences of the primers used to validate CNVRs by qPCR. (XLSX 9 kb)
Data Availability Statement
Microarray expression data can be found in 10.6084/m9.figshare.7952054, while Porcine SNP60 BeadChip data can be accessed at 10.6084/m9.figshare.7473092.v2