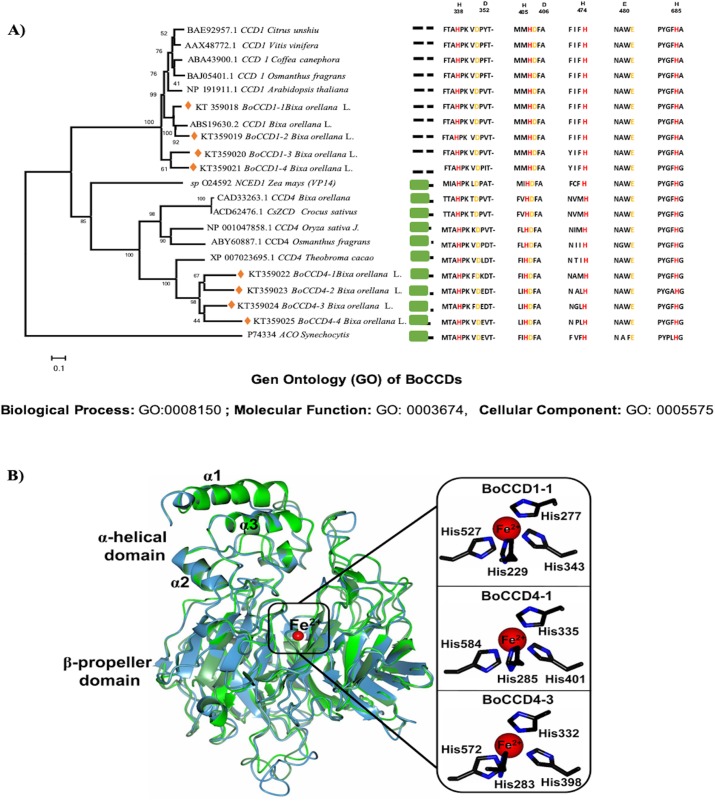
Figure 3. In silico analysis of BoCCDs.
(A) Sequence alignments of CCD enzymes, alignment of B. orellana BoCCD-like proteins and subcellular locations of BoCCDs by iPSORT. The phylogenetic tree was inferred using the maximum-likelihood method based on the Jones–Taylor–Thornton (JTT) substitution model and was gamma distributed with Invariant sites (G + I) in MEGA6. Osmanthus fragrans (ABY60887.1), Oryza sativa Japonica group (NP_001047858.1), Crocus sativus (ACD62476.1), Theobroma cacao (XP_007023695.1), Z. mays (O24592), Citrus unshiu (BAE92957.1), Vitis vinifera (AAX48772.1), Coffea canephora (ABA43900.1), O. fragrans (BAJ05401.1), B. orellana L. (CAD71148.1, ABS19630.2, KT359018, KT359019, KT359020, KT359021, KT359022, KT359023, KT359024), and Arabidopsis thaliana (NP_191911.1). Numbers near the branch points represent the bootstrap value produced by 1,000 replications. Phylogenetic trees were rooted with Synechocystis apocarotenoid cleavage oxygenase (ACO) (P74334). Orange diamonds in the tree indicate the sequences in the study. The representative alignment indicates the signal peptide in the green rectangle according to iPSORT. Red letters indicate residues of histidine, and yellow letters indicate residues of aspartate or glutamate. Gene ontology (GO) annotation was performed with Blast2GO software for InterPro scanning to determine potential function of BoCCDs. Top 10 GO description in the three main categories, biological process, molecular function, and cellular component (See Dataset3, Götz et al., 2008). (B) Structural superposition of BoCCD1-1 (green), BoCCD4-1 (light blue), and BoCCD4-3 (lawn green) homology models showing the catalytic iron (red sphere) and α-helical and β-propeller domains. Right inset, close-up of four Fe2+-coordinating histidine residues (black cylinders).