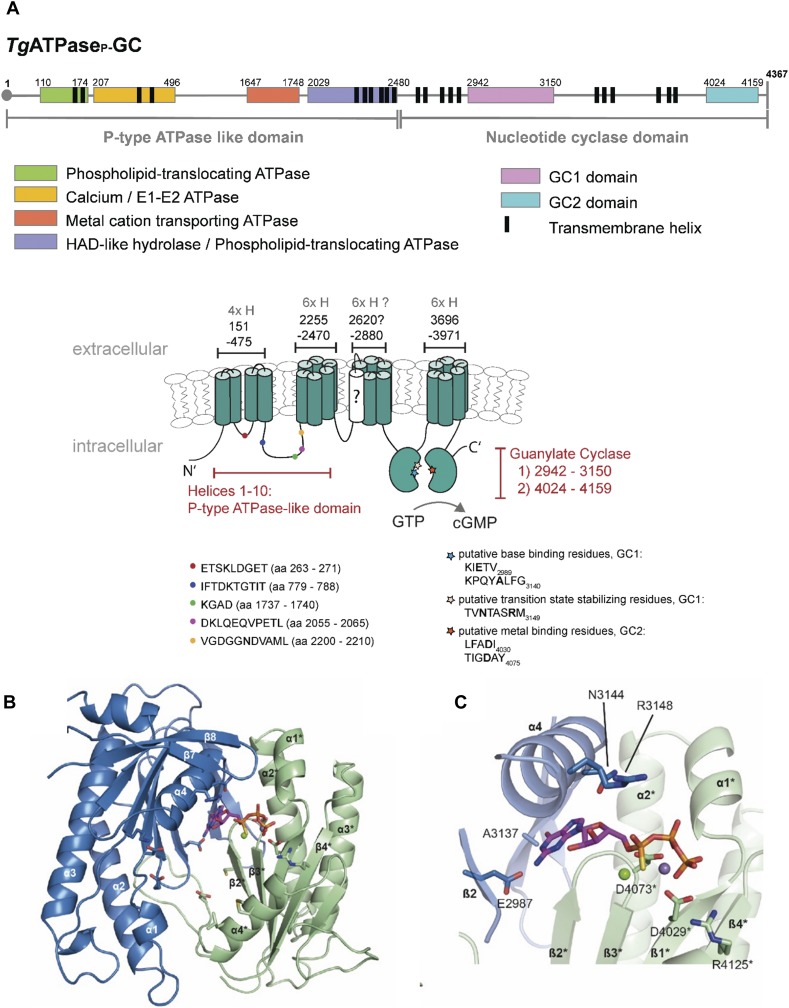
Figure 1. The genome of T. gondii harbors an unusual heterodimeric GC conjugated to P-type ATPase domain.
(A) The primary and secondary topology of TgATPaseP-GC as predicted using TMHMM, SMART, TMpred, Phobius, and NCBI domain search tools. The model was constructed by consensus across algorithms regarding the position of domains and transmembrane spans. The N terminus (1–2,480 aa) containing 10 α-helices resembles P-type ATPase with at least four subdomains (color-coded). The C terminus (2,481–4,367 aa) harbors two potential nucleotide cyclase catalytic regions, termed GC1 and GC2, each following six transmembrane helices. The question-marked (?) helix was predicted only by Phobius (probability score, 752). The color-coded signs on secondary structure show the position of highly conserved sequences in the ATPase and cyclase domains. The key residues involved in the base binding and catalysis of cyclases are also depicted in bold letters. (B, C) Tertiary structure of GC1 and GC2 domains based on homology modeling. The ribbon diagrams of GC1 and GC2 suggest a functional activation by pseudo-heterodimerization similar to tmAC. The model shows an antiparallel arrangement of GC1 and GC2, where each domain harbors a seven-stranded β-sheet surrounded by three α-helices. The image in panel (C) illustrates a GC1-GC2 heterodimer interface bound to GTPαS. The residues of GC2 labeled with asterisk (*) interact with the phosphate backbone of the nucleotide.